
Operophtera brumata reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; unclassified Reoviridae
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
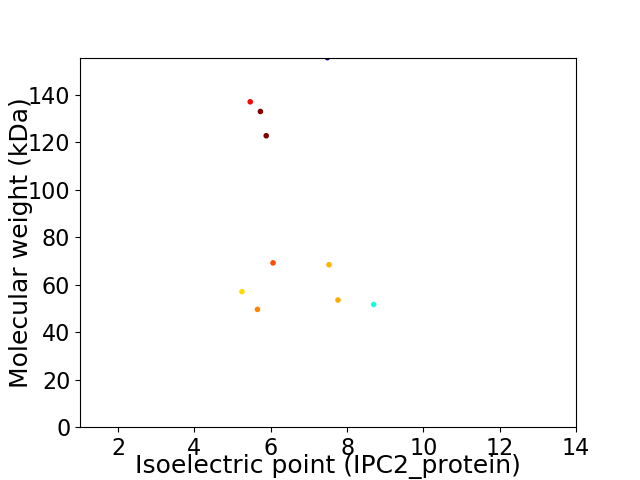
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q30C80|Q30C80_9REOV Uncharacterized protein OS=Operophtera brumata reovirus OX=352248 PE=4 SV=1
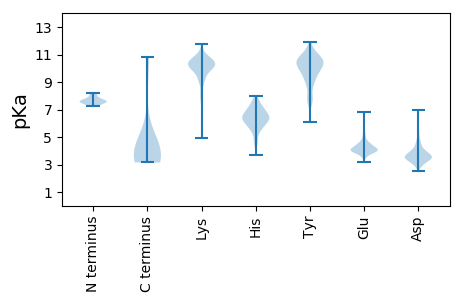
MM1 pKa = 7.42EE2 pKa = 5.65RR3 pKa = 11.84LINSYY8 pKa = 8.75RR9 pKa = 11.84TISHH13 pKa = 6.6SLRR16 pKa = 11.84QLVPAYY22 pKa = 7.18TTFEE26 pKa = 4.28DD27 pKa = 5.8LEE29 pKa = 4.45AQLPPDD35 pKa = 3.59YY36 pKa = 10.94VFPEE40 pKa = 4.25SFQYY44 pKa = 9.99PPRR47 pKa = 11.84IRR49 pKa = 11.84YY50 pKa = 9.18RR51 pKa = 11.84EE52 pKa = 3.87IHH54 pKa = 6.41SDD56 pKa = 2.94AGFTRR61 pKa = 11.84EE62 pKa = 4.25VEE64 pKa = 3.72AAMAGRR70 pKa = 11.84LANDD74 pKa = 3.46EE75 pKa = 4.98DD76 pKa = 4.15NPIMFDD82 pKa = 3.03YY83 pKa = 10.67RR84 pKa = 11.84EE85 pKa = 3.97PVRR88 pKa = 11.84PGRR91 pKa = 11.84IANWLTGYY99 pKa = 11.05AEE101 pKa = 4.3AANTLFDD108 pKa = 3.79QVLSEE113 pKa = 4.12VDD115 pKa = 3.07ALASVFYY122 pKa = 10.93DD123 pKa = 2.85VDD125 pKa = 3.33EE126 pKa = 4.28LLRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84FQEE134 pKa = 3.92GDD136 pKa = 3.88VIPNDD141 pKa = 3.79HH142 pKa = 7.25DD143 pKa = 4.2HH144 pKa = 7.37PSNDD148 pKa = 2.97VLEE151 pKa = 4.71PSEE154 pKa = 4.85HH155 pKa = 5.6EE156 pKa = 3.99AARR159 pKa = 11.84VHH161 pKa = 6.53SDD163 pKa = 3.06QVVEE167 pKa = 4.24EE168 pKa = 4.29RR169 pKa = 11.84VQRR172 pKa = 11.84PLVRR176 pKa = 11.84LEE178 pKa = 4.18SPGEE182 pKa = 3.73DD183 pKa = 2.98EE184 pKa = 5.32IIFRR188 pKa = 11.84GPSDD192 pKa = 3.13IRR194 pKa = 11.84YY195 pKa = 7.72TPITLRR201 pKa = 11.84THH203 pKa = 6.24FHH205 pKa = 6.59FKK207 pKa = 9.5TFEE210 pKa = 4.04RR211 pKa = 11.84PVEE214 pKa = 4.02KK215 pKa = 10.45DD216 pKa = 2.95QPYY219 pKa = 10.65PSFKK223 pKa = 10.77VNQLHH228 pKa = 7.16PNTGLDD234 pKa = 3.51FSGRR238 pKa = 11.84PISSSFISAFNMSINPEE255 pKa = 4.15LASVTSALVYY265 pKa = 10.48LLNEE269 pKa = 3.48GRR271 pKa = 11.84YY272 pKa = 9.25GVRR275 pKa = 11.84DD276 pKa = 3.67DD277 pKa = 4.25SLVYY281 pKa = 10.41SHH283 pKa = 6.64NTTHH287 pKa = 6.12NQISPASTKK296 pKa = 10.5AALKK300 pKa = 9.27MEE302 pKa = 4.0EE303 pKa = 4.08RR304 pKa = 11.84VRR306 pKa = 11.84DD307 pKa = 3.67SVTYY311 pKa = 9.99FSSKK315 pKa = 10.16RR316 pKa = 11.84LTRR319 pKa = 11.84GQCEE323 pKa = 3.59HH324 pKa = 7.72LIRR327 pKa = 11.84HH328 pKa = 6.23MISYY332 pKa = 9.85HH333 pKa = 6.02INTPDD338 pKa = 3.01THH340 pKa = 7.81GLIVEE345 pKa = 4.84CYY347 pKa = 8.97EE348 pKa = 4.05QVGDD352 pKa = 4.07LRR354 pKa = 11.84ADD356 pKa = 3.62FTLEE360 pKa = 3.82SLRR363 pKa = 11.84IPLLNVDD370 pKa = 3.71WTRR373 pKa = 11.84ICYY376 pKa = 10.11NISNALEE383 pKa = 4.25NEE385 pKa = 4.11LEE387 pKa = 3.97RR388 pKa = 11.84AIFDD392 pKa = 3.25VLMFVAIMKK401 pKa = 9.21FSVVVTEE408 pKa = 3.96PQRR411 pKa = 11.84MVDD414 pKa = 3.55IAPGPFAVSAFLGCVPGSVVTHH436 pKa = 6.85DD437 pKa = 3.64AFKK440 pKa = 10.6TEE442 pKa = 3.86WKK444 pKa = 9.08RR445 pKa = 11.84TISRR449 pKa = 11.84PNSSTEE455 pKa = 3.48WPLTKK460 pKa = 9.97TSYY463 pKa = 10.54HH464 pKa = 6.1FPQAIGVPCTQVMPITRR481 pKa = 11.84RR482 pKa = 11.84PDD484 pKa = 3.23FQMFGCLLSMVINNMM499 pKa = 3.49
MM1 pKa = 7.42EE2 pKa = 5.65RR3 pKa = 11.84LINSYY8 pKa = 8.75RR9 pKa = 11.84TISHH13 pKa = 6.6SLRR16 pKa = 11.84QLVPAYY22 pKa = 7.18TTFEE26 pKa = 4.28DD27 pKa = 5.8LEE29 pKa = 4.45AQLPPDD35 pKa = 3.59YY36 pKa = 10.94VFPEE40 pKa = 4.25SFQYY44 pKa = 9.99PPRR47 pKa = 11.84IRR49 pKa = 11.84YY50 pKa = 9.18RR51 pKa = 11.84EE52 pKa = 3.87IHH54 pKa = 6.41SDD56 pKa = 2.94AGFTRR61 pKa = 11.84EE62 pKa = 4.25VEE64 pKa = 3.72AAMAGRR70 pKa = 11.84LANDD74 pKa = 3.46EE75 pKa = 4.98DD76 pKa = 4.15NPIMFDD82 pKa = 3.03YY83 pKa = 10.67RR84 pKa = 11.84EE85 pKa = 3.97PVRR88 pKa = 11.84PGRR91 pKa = 11.84IANWLTGYY99 pKa = 11.05AEE101 pKa = 4.3AANTLFDD108 pKa = 3.79QVLSEE113 pKa = 4.12VDD115 pKa = 3.07ALASVFYY122 pKa = 10.93DD123 pKa = 2.85VDD125 pKa = 3.33EE126 pKa = 4.28LLRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84FQEE134 pKa = 3.92GDD136 pKa = 3.88VIPNDD141 pKa = 3.79HH142 pKa = 7.25DD143 pKa = 4.2HH144 pKa = 7.37PSNDD148 pKa = 2.97VLEE151 pKa = 4.71PSEE154 pKa = 4.85HH155 pKa = 5.6EE156 pKa = 3.99AARR159 pKa = 11.84VHH161 pKa = 6.53SDD163 pKa = 3.06QVVEE167 pKa = 4.24EE168 pKa = 4.29RR169 pKa = 11.84VQRR172 pKa = 11.84PLVRR176 pKa = 11.84LEE178 pKa = 4.18SPGEE182 pKa = 3.73DD183 pKa = 2.98EE184 pKa = 5.32IIFRR188 pKa = 11.84GPSDD192 pKa = 3.13IRR194 pKa = 11.84YY195 pKa = 7.72TPITLRR201 pKa = 11.84THH203 pKa = 6.24FHH205 pKa = 6.59FKK207 pKa = 9.5TFEE210 pKa = 4.04RR211 pKa = 11.84PVEE214 pKa = 4.02KK215 pKa = 10.45DD216 pKa = 2.95QPYY219 pKa = 10.65PSFKK223 pKa = 10.77VNQLHH228 pKa = 7.16PNTGLDD234 pKa = 3.51FSGRR238 pKa = 11.84PISSSFISAFNMSINPEE255 pKa = 4.15LASVTSALVYY265 pKa = 10.48LLNEE269 pKa = 3.48GRR271 pKa = 11.84YY272 pKa = 9.25GVRR275 pKa = 11.84DD276 pKa = 3.67DD277 pKa = 4.25SLVYY281 pKa = 10.41SHH283 pKa = 6.64NTTHH287 pKa = 6.12NQISPASTKK296 pKa = 10.5AALKK300 pKa = 9.27MEE302 pKa = 4.0EE303 pKa = 4.08RR304 pKa = 11.84VRR306 pKa = 11.84DD307 pKa = 3.67SVTYY311 pKa = 9.99FSSKK315 pKa = 10.16RR316 pKa = 11.84LTRR319 pKa = 11.84GQCEE323 pKa = 3.59HH324 pKa = 7.72LIRR327 pKa = 11.84HH328 pKa = 6.23MISYY332 pKa = 9.85HH333 pKa = 6.02INTPDD338 pKa = 3.01THH340 pKa = 7.81GLIVEE345 pKa = 4.84CYY347 pKa = 8.97EE348 pKa = 4.05QVGDD352 pKa = 4.07LRR354 pKa = 11.84ADD356 pKa = 3.62FTLEE360 pKa = 3.82SLRR363 pKa = 11.84IPLLNVDD370 pKa = 3.71WTRR373 pKa = 11.84ICYY376 pKa = 10.11NISNALEE383 pKa = 4.25NEE385 pKa = 4.11LEE387 pKa = 3.97RR388 pKa = 11.84AIFDD392 pKa = 3.25VLMFVAIMKK401 pKa = 9.21FSVVVTEE408 pKa = 3.96PQRR411 pKa = 11.84MVDD414 pKa = 3.55IAPGPFAVSAFLGCVPGSVVTHH436 pKa = 6.85DD437 pKa = 3.64AFKK440 pKa = 10.6TEE442 pKa = 3.86WKK444 pKa = 9.08RR445 pKa = 11.84TISRR449 pKa = 11.84PNSSTEE455 pKa = 3.48WPLTKK460 pKa = 9.97TSYY463 pKa = 10.54HH464 pKa = 6.1FPQAIGVPCTQVMPITRR481 pKa = 11.84RR482 pKa = 11.84PDD484 pKa = 3.23FQMFGCLLSMVINNMM499 pKa = 3.49
Molecular weight: 57.1 kDa
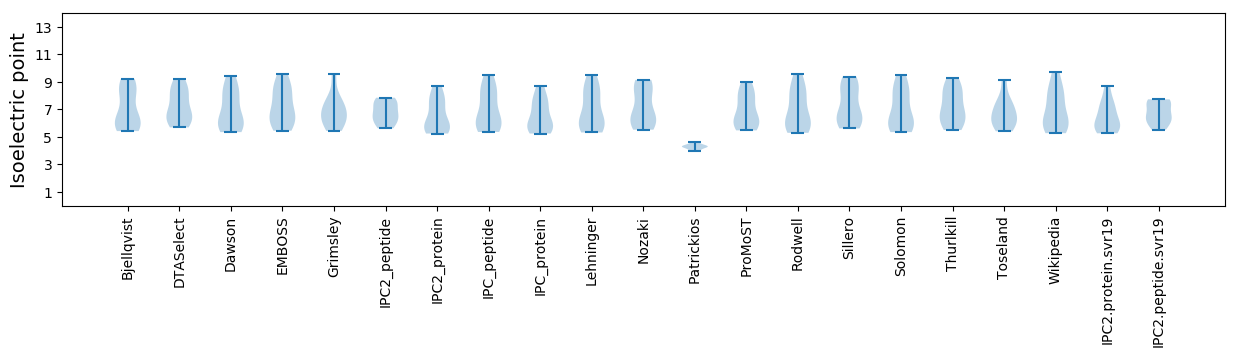
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q30C79|Q30C79_9REOV Uncharacterized protein OS=Operophtera brumata reovirus OX=352248 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 5.49NDD4 pKa = 4.08SYY6 pKa = 11.78LSDD9 pKa = 3.68LTPFVGPVSYY19 pKa = 10.64VGRR22 pKa = 11.84NLTRR26 pKa = 11.84ILYY29 pKa = 9.38DD30 pKa = 3.76LQALSSKK37 pKa = 10.49CRR39 pKa = 11.84GKK41 pKa = 10.74VLLSSKK47 pKa = 9.17TGQPPRR53 pKa = 11.84VQVTRR58 pKa = 11.84ILVEE62 pKa = 3.86QMFTVYY68 pKa = 9.22TQLRR72 pKa = 11.84GYY74 pKa = 9.57NFSVTLTSLLNEE86 pKa = 4.76LSTALAMSVGRR97 pKa = 11.84SEE99 pKa = 4.33LTLNSNVLYY108 pKa = 9.98MITNVNSDD116 pKa = 2.96IAISIGKK123 pKa = 8.13VWNGVPPLRR132 pKa = 11.84LLNGSGSNADD142 pKa = 3.28VTIIGDD148 pKa = 3.93DD149 pKa = 3.97NILTRR154 pKa = 11.84MLRR157 pKa = 11.84KK158 pKa = 9.84LPGEE162 pKa = 4.0ARR164 pKa = 11.84VEE166 pKa = 4.07LTKK169 pKa = 10.64LAQVSGQLAGVRR181 pKa = 11.84VVRR184 pKa = 11.84QKK186 pKa = 11.45DD187 pKa = 3.68VTTLLADD194 pKa = 3.48QLFSRR199 pKa = 11.84SSGSIDD205 pKa = 3.39LARR208 pKa = 11.84IFVQMNKK215 pKa = 9.19QATVLSQFKK224 pKa = 9.97TDD226 pKa = 4.57FLMTPLSRR234 pKa = 11.84FHH236 pKa = 7.44DD237 pKa = 3.99LGRR240 pKa = 11.84PLFIEE245 pKa = 4.33IDD247 pKa = 3.87LVLTKK252 pKa = 10.48ADD254 pKa = 3.85SNSILSAPSISGEE267 pKa = 3.96VLFTIRR273 pKa = 11.84STADD277 pKa = 2.99VEE279 pKa = 4.51LSFSILRR286 pKa = 11.84GEE288 pKa = 4.06TSTIIKK294 pKa = 8.25LQPYY298 pKa = 9.13SHH300 pKa = 5.76VQRR303 pKa = 11.84FGEE306 pKa = 4.37KK307 pKa = 7.86VTFPTGKK314 pKa = 10.52GGILVTATGDD324 pKa = 3.28LRR326 pKa = 11.84NSVRR330 pKa = 11.84SGQRR334 pKa = 11.84SLIEE338 pKa = 3.83MGIEE342 pKa = 4.01GTLGQTTNTEE352 pKa = 3.94LSRR355 pKa = 11.84WFYY358 pKa = 11.03QIVTPILPYY367 pKa = 10.79LPGIVSMLTTSQPFNSVTRR386 pKa = 11.84VISPEE391 pKa = 3.51VSLYY395 pKa = 9.69FQNLKK400 pKa = 10.43PSFYY404 pKa = 10.76EE405 pKa = 4.17DD406 pKa = 3.98LLTTGTTPADD416 pKa = 3.32KK417 pKa = 10.35QRR419 pKa = 11.84RR420 pKa = 11.84VLVAYY425 pKa = 7.58TLILLIPEE433 pKa = 4.13YY434 pKa = 10.67LLNPVITLNNVNEE447 pKa = 4.51TPPFMSMYY455 pKa = 9.73GALVGHH461 pKa = 6.5FRR463 pKa = 11.84QIYY466 pKa = 7.59EE467 pKa = 3.89
MM1 pKa = 7.63EE2 pKa = 5.49NDD4 pKa = 4.08SYY6 pKa = 11.78LSDD9 pKa = 3.68LTPFVGPVSYY19 pKa = 10.64VGRR22 pKa = 11.84NLTRR26 pKa = 11.84ILYY29 pKa = 9.38DD30 pKa = 3.76LQALSSKK37 pKa = 10.49CRR39 pKa = 11.84GKK41 pKa = 10.74VLLSSKK47 pKa = 9.17TGQPPRR53 pKa = 11.84VQVTRR58 pKa = 11.84ILVEE62 pKa = 3.86QMFTVYY68 pKa = 9.22TQLRR72 pKa = 11.84GYY74 pKa = 9.57NFSVTLTSLLNEE86 pKa = 4.76LSTALAMSVGRR97 pKa = 11.84SEE99 pKa = 4.33LTLNSNVLYY108 pKa = 9.98MITNVNSDD116 pKa = 2.96IAISIGKK123 pKa = 8.13VWNGVPPLRR132 pKa = 11.84LLNGSGSNADD142 pKa = 3.28VTIIGDD148 pKa = 3.93DD149 pKa = 3.97NILTRR154 pKa = 11.84MLRR157 pKa = 11.84KK158 pKa = 9.84LPGEE162 pKa = 4.0ARR164 pKa = 11.84VEE166 pKa = 4.07LTKK169 pKa = 10.64LAQVSGQLAGVRR181 pKa = 11.84VVRR184 pKa = 11.84QKK186 pKa = 11.45DD187 pKa = 3.68VTTLLADD194 pKa = 3.48QLFSRR199 pKa = 11.84SSGSIDD205 pKa = 3.39LARR208 pKa = 11.84IFVQMNKK215 pKa = 9.19QATVLSQFKK224 pKa = 9.97TDD226 pKa = 4.57FLMTPLSRR234 pKa = 11.84FHH236 pKa = 7.44DD237 pKa = 3.99LGRR240 pKa = 11.84PLFIEE245 pKa = 4.33IDD247 pKa = 3.87LVLTKK252 pKa = 10.48ADD254 pKa = 3.85SNSILSAPSISGEE267 pKa = 3.96VLFTIRR273 pKa = 11.84STADD277 pKa = 2.99VEE279 pKa = 4.51LSFSILRR286 pKa = 11.84GEE288 pKa = 4.06TSTIIKK294 pKa = 8.25LQPYY298 pKa = 9.13SHH300 pKa = 5.76VQRR303 pKa = 11.84FGEE306 pKa = 4.37KK307 pKa = 7.86VTFPTGKK314 pKa = 10.52GGILVTATGDD324 pKa = 3.28LRR326 pKa = 11.84NSVRR330 pKa = 11.84SGQRR334 pKa = 11.84SLIEE338 pKa = 3.83MGIEE342 pKa = 4.01GTLGQTTNTEE352 pKa = 3.94LSRR355 pKa = 11.84WFYY358 pKa = 11.03QIVTPILPYY367 pKa = 10.79LPGIVSMLTTSQPFNSVTRR386 pKa = 11.84VISPEE391 pKa = 3.51VSLYY395 pKa = 9.69FQNLKK400 pKa = 10.43PSFYY404 pKa = 10.76EE405 pKa = 4.17DD406 pKa = 3.98LLTTGTTPADD416 pKa = 3.32KK417 pKa = 10.35QRR419 pKa = 11.84RR420 pKa = 11.84VLVAYY425 pKa = 7.58TLILLIPEE433 pKa = 4.13YY434 pKa = 10.67LLNPVITLNNVNEE447 pKa = 4.51TPPFMSMYY455 pKa = 9.73GALVGHH461 pKa = 6.5FRR463 pKa = 11.84QIYY466 pKa = 7.59EE467 pKa = 3.89
Molecular weight: 51.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7901 |
437 |
1358 |
790.1 |
89.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.404 ± 0.342 | 0.899 ± 0.174 |
5.594 ± 0.233 | 5.442 ± 0.256 |
4.949 ± 0.311 | 4.999 ± 0.186 |
2.493 ± 0.298 | 6.404 ± 0.264 |
3.721 ± 0.237 | 10.543 ± 0.523 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.569 ± 0.177 | 4.303 ± 0.171 |
4.772 ± 0.263 | 4.354 ± 0.283 |
6.683 ± 0.174 | 7.543 ± 0.422 |
7.771 ± 0.416 | 7.1 ± 0.25 |
0.987 ± 0.105 | 3.468 ± 0.131 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
