
Capybara microvirus Cap1_SP_151
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
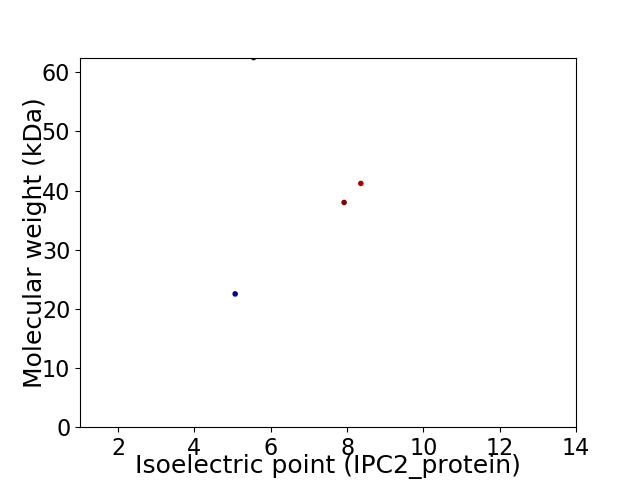
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVQ1|A0A4V1FVQ1_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_151 OX=2585393 PE=4 SV=1
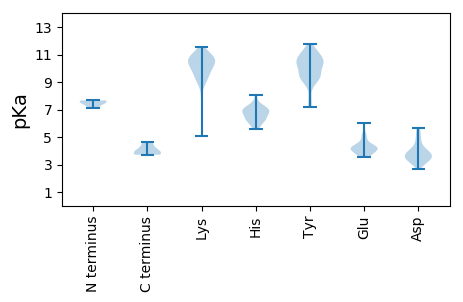
MM1 pKa = 7.66IRR3 pKa = 11.84RR4 pKa = 11.84FWMRR8 pKa = 11.84KK9 pKa = 6.7EE10 pKa = 4.84LIDD13 pKa = 5.4SVEE16 pKa = 4.13TDD18 pKa = 2.82QNTAFRR24 pKa = 11.84IKK26 pKa = 9.78TKK28 pKa = 10.59FNFIVYY34 pKa = 9.02PSNSGSKK41 pKa = 9.32FHH43 pKa = 6.59TKK45 pKa = 10.37YY46 pKa = 10.37EE47 pKa = 4.23YY48 pKa = 9.41KK49 pKa = 7.74TTQGITEE56 pKa = 4.5LVPTEE61 pKa = 4.02EE62 pKa = 4.19VNTYY66 pKa = 11.16NEE68 pKa = 4.04VQSYY72 pKa = 10.02KK73 pKa = 10.75EE74 pKa = 3.65EE75 pKa = 3.87TSIYY79 pKa = 9.98KK80 pKa = 10.12ILDD83 pKa = 2.96RR84 pKa = 11.84MTRR87 pKa = 11.84GDD89 pKa = 4.03YY90 pKa = 10.75SALDD94 pKa = 3.63KK95 pKa = 11.39NQGIFMDD102 pKa = 4.66CTNLPKK108 pKa = 10.49DD109 pKa = 3.61INEE112 pKa = 4.1LTDD115 pKa = 3.46FFNNYY120 pKa = 9.11EE121 pKa = 4.28STFNQADD128 pKa = 3.53PRR130 pKa = 11.84FKK132 pKa = 10.97EE133 pKa = 4.17LFNNNYY139 pKa = 8.95KK140 pKa = 10.17QLIFDD145 pKa = 4.36FQNGTFEE152 pKa = 4.48EE153 pKa = 5.09KK154 pKa = 9.65YY155 pKa = 10.36KK156 pKa = 10.74KK157 pKa = 10.53YY158 pKa = 10.84NEE160 pKa = 4.4LYY162 pKa = 10.68SPEE165 pKa = 4.15KK166 pKa = 10.11LQSNNVVSSEE176 pKa = 4.32PINFEE181 pKa = 3.92NKK183 pKa = 8.97EE184 pKa = 4.14VNDD187 pKa = 3.78NGG189 pKa = 3.92
MM1 pKa = 7.66IRR3 pKa = 11.84RR4 pKa = 11.84FWMRR8 pKa = 11.84KK9 pKa = 6.7EE10 pKa = 4.84LIDD13 pKa = 5.4SVEE16 pKa = 4.13TDD18 pKa = 2.82QNTAFRR24 pKa = 11.84IKK26 pKa = 9.78TKK28 pKa = 10.59FNFIVYY34 pKa = 9.02PSNSGSKK41 pKa = 9.32FHH43 pKa = 6.59TKK45 pKa = 10.37YY46 pKa = 10.37EE47 pKa = 4.23YY48 pKa = 9.41KK49 pKa = 7.74TTQGITEE56 pKa = 4.5LVPTEE61 pKa = 4.02EE62 pKa = 4.19VNTYY66 pKa = 11.16NEE68 pKa = 4.04VQSYY72 pKa = 10.02KK73 pKa = 10.75EE74 pKa = 3.65EE75 pKa = 3.87TSIYY79 pKa = 9.98KK80 pKa = 10.12ILDD83 pKa = 2.96RR84 pKa = 11.84MTRR87 pKa = 11.84GDD89 pKa = 4.03YY90 pKa = 10.75SALDD94 pKa = 3.63KK95 pKa = 11.39NQGIFMDD102 pKa = 4.66CTNLPKK108 pKa = 10.49DD109 pKa = 3.61INEE112 pKa = 4.1LTDD115 pKa = 3.46FFNNYY120 pKa = 9.11EE121 pKa = 4.28STFNQADD128 pKa = 3.53PRR130 pKa = 11.84FKK132 pKa = 10.97EE133 pKa = 4.17LFNNNYY139 pKa = 8.95KK140 pKa = 10.17QLIFDD145 pKa = 4.36FQNGTFEE152 pKa = 4.48EE153 pKa = 5.09KK154 pKa = 9.65YY155 pKa = 10.36KK156 pKa = 10.74KK157 pKa = 10.53YY158 pKa = 10.84NEE160 pKa = 4.4LYY162 pKa = 10.68SPEE165 pKa = 4.15KK166 pKa = 10.11LQSNNVVSSEE176 pKa = 4.32PINFEE181 pKa = 3.92NKK183 pKa = 8.97EE184 pKa = 4.14VNDD187 pKa = 3.78NGG189 pKa = 3.92
Molecular weight: 22.53 kDa
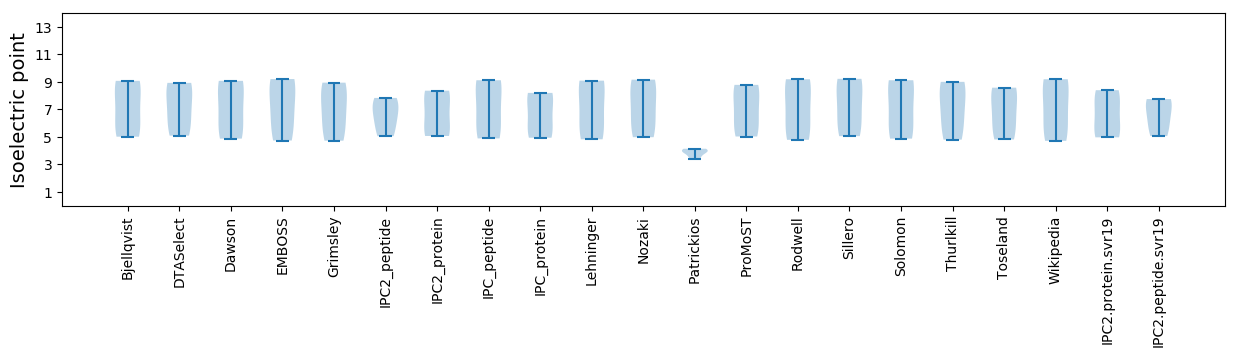
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W590|A0A4P8W590_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_151 OX=2585393 PE=4 SV=1
MM1 pKa = 7.1QCFDD5 pKa = 4.5PFVVSNIRR13 pKa = 11.84NPLIQLNYY21 pKa = 9.19KK22 pKa = 9.93ASKK25 pKa = 9.67KK26 pKa = 10.13RR27 pKa = 11.84RR28 pKa = 11.84FAYY31 pKa = 9.11WIKK34 pKa = 10.75FKK36 pKa = 10.72DD37 pKa = 3.28IRR39 pKa = 11.84KK40 pKa = 9.47DD41 pKa = 3.38LLPEE45 pKa = 3.59VLEE48 pKa = 4.54RR49 pKa = 11.84IKK51 pKa = 11.26NKK53 pKa = 10.58DD54 pKa = 3.32MLLIPCGRR62 pKa = 11.84CFSCRR67 pKa = 11.84SKK69 pKa = 11.14KK70 pKa = 10.45AMSWTIRR77 pKa = 11.84NYY79 pKa = 10.29FEE81 pKa = 4.71SEE83 pKa = 4.28KK84 pKa = 10.71YY85 pKa = 9.82DD86 pKa = 3.34QKK88 pKa = 11.52CFITATYY95 pKa = 11.16NEE97 pKa = 4.63FKK99 pKa = 10.74CPRR102 pKa = 11.84FLVKK106 pKa = 10.1KK107 pKa = 10.28DD108 pKa = 3.42LQLLIKK114 pKa = 9.82QIRR117 pKa = 11.84RR118 pKa = 11.84YY119 pKa = 9.3IDD121 pKa = 3.02YY122 pKa = 10.35HH123 pKa = 8.03ISSDD127 pKa = 3.45MKK129 pKa = 10.42IKK131 pKa = 10.8YY132 pKa = 7.35FACGEE137 pKa = 4.04YY138 pKa = 10.37GGQFGRR144 pKa = 11.84PHH146 pKa = 5.83FHH148 pKa = 6.36ILLYY152 pKa = 10.19GFCFPKK158 pKa = 10.7DD159 pKa = 3.69HH160 pKa = 6.93ILKK163 pKa = 10.59DD164 pKa = 3.73EE165 pKa = 4.39NGKK168 pKa = 9.79KK169 pKa = 10.09EE170 pKa = 4.15YY171 pKa = 10.1ISLKK175 pKa = 9.74LTEE178 pKa = 4.18LWNRR182 pKa = 11.84GIISFSDD189 pKa = 3.86CVDD192 pKa = 3.3ASHH195 pKa = 7.42IKK197 pKa = 8.0YY198 pKa = 8.01TCGYY202 pKa = 7.17VQKK205 pKa = 10.43KK206 pKa = 9.68LFHH209 pKa = 6.9IYY211 pKa = 10.58SDD213 pKa = 3.92AEE215 pKa = 4.18KK216 pKa = 10.89KK217 pKa = 10.55LLQDD221 pKa = 4.15WIEE224 pKa = 4.04LHH226 pKa = 6.96PDD228 pKa = 3.01DD229 pKa = 5.63HH230 pKa = 6.91FQDD233 pKa = 4.29SFITCSKK240 pKa = 10.86GIGLEE245 pKa = 4.04FFDD248 pKa = 3.75KK249 pKa = 10.97NKK251 pKa = 10.66YY252 pKa = 9.92NIYY255 pKa = 10.43NDD257 pKa = 3.48NEE259 pKa = 4.36FLFHH263 pKa = 7.15PPFDD267 pKa = 4.12SSPVKK272 pKa = 10.06HH273 pKa = 6.22VKK275 pKa = 9.34PLPYY279 pKa = 9.47FDD281 pKa = 5.44YY282 pKa = 11.14RR283 pKa = 11.84MQNVDD288 pKa = 3.52YY289 pKa = 11.01NLYY292 pKa = 10.3AHH294 pKa = 6.78SKK296 pKa = 9.58FLRR299 pKa = 11.84SEE301 pKa = 4.12YY302 pKa = 10.75NEE304 pKa = 3.84NYY306 pKa = 9.51YY307 pKa = 11.04KK308 pKa = 10.81KK309 pKa = 10.35FLEE312 pKa = 4.37DD313 pKa = 3.58DD314 pKa = 3.73RR315 pKa = 11.84SISEE319 pKa = 3.81IMRR322 pKa = 11.84AEE324 pKa = 3.76EE325 pKa = 5.29DD326 pKa = 3.88YY327 pKa = 11.58ALKK330 pKa = 10.56NYY332 pKa = 10.37KK333 pKa = 9.73LYY335 pKa = 10.84KK336 pKa = 9.06YY337 pKa = 10.08IRR339 pKa = 11.84RR340 pKa = 3.86
MM1 pKa = 7.1QCFDD5 pKa = 4.5PFVVSNIRR13 pKa = 11.84NPLIQLNYY21 pKa = 9.19KK22 pKa = 9.93ASKK25 pKa = 9.67KK26 pKa = 10.13RR27 pKa = 11.84RR28 pKa = 11.84FAYY31 pKa = 9.11WIKK34 pKa = 10.75FKK36 pKa = 10.72DD37 pKa = 3.28IRR39 pKa = 11.84KK40 pKa = 9.47DD41 pKa = 3.38LLPEE45 pKa = 3.59VLEE48 pKa = 4.54RR49 pKa = 11.84IKK51 pKa = 11.26NKK53 pKa = 10.58DD54 pKa = 3.32MLLIPCGRR62 pKa = 11.84CFSCRR67 pKa = 11.84SKK69 pKa = 11.14KK70 pKa = 10.45AMSWTIRR77 pKa = 11.84NYY79 pKa = 10.29FEE81 pKa = 4.71SEE83 pKa = 4.28KK84 pKa = 10.71YY85 pKa = 9.82DD86 pKa = 3.34QKK88 pKa = 11.52CFITATYY95 pKa = 11.16NEE97 pKa = 4.63FKK99 pKa = 10.74CPRR102 pKa = 11.84FLVKK106 pKa = 10.1KK107 pKa = 10.28DD108 pKa = 3.42LQLLIKK114 pKa = 9.82QIRR117 pKa = 11.84RR118 pKa = 11.84YY119 pKa = 9.3IDD121 pKa = 3.02YY122 pKa = 10.35HH123 pKa = 8.03ISSDD127 pKa = 3.45MKK129 pKa = 10.42IKK131 pKa = 10.8YY132 pKa = 7.35FACGEE137 pKa = 4.04YY138 pKa = 10.37GGQFGRR144 pKa = 11.84PHH146 pKa = 5.83FHH148 pKa = 6.36ILLYY152 pKa = 10.19GFCFPKK158 pKa = 10.7DD159 pKa = 3.69HH160 pKa = 6.93ILKK163 pKa = 10.59DD164 pKa = 3.73EE165 pKa = 4.39NGKK168 pKa = 9.79KK169 pKa = 10.09EE170 pKa = 4.15YY171 pKa = 10.1ISLKK175 pKa = 9.74LTEE178 pKa = 4.18LWNRR182 pKa = 11.84GIISFSDD189 pKa = 3.86CVDD192 pKa = 3.3ASHH195 pKa = 7.42IKK197 pKa = 8.0YY198 pKa = 8.01TCGYY202 pKa = 7.17VQKK205 pKa = 10.43KK206 pKa = 9.68LFHH209 pKa = 6.9IYY211 pKa = 10.58SDD213 pKa = 3.92AEE215 pKa = 4.18KK216 pKa = 10.89KK217 pKa = 10.55LLQDD221 pKa = 4.15WIEE224 pKa = 4.04LHH226 pKa = 6.96PDD228 pKa = 3.01DD229 pKa = 5.63HH230 pKa = 6.91FQDD233 pKa = 4.29SFITCSKK240 pKa = 10.86GIGLEE245 pKa = 4.04FFDD248 pKa = 3.75KK249 pKa = 10.97NKK251 pKa = 10.66YY252 pKa = 9.92NIYY255 pKa = 10.43NDD257 pKa = 3.48NEE259 pKa = 4.36FLFHH263 pKa = 7.15PPFDD267 pKa = 4.12SSPVKK272 pKa = 10.06HH273 pKa = 6.22VKK275 pKa = 9.34PLPYY279 pKa = 9.47FDD281 pKa = 5.44YY282 pKa = 11.14RR283 pKa = 11.84MQNVDD288 pKa = 3.52YY289 pKa = 11.01NLYY292 pKa = 10.3AHH294 pKa = 6.78SKK296 pKa = 9.58FLRR299 pKa = 11.84SEE301 pKa = 4.12YY302 pKa = 10.75NEE304 pKa = 3.84NYY306 pKa = 9.51YY307 pKa = 11.04KK308 pKa = 10.81KK309 pKa = 10.35FLEE312 pKa = 4.37DD313 pKa = 3.58DD314 pKa = 3.73RR315 pKa = 11.84SISEE319 pKa = 3.81IMRR322 pKa = 11.84AEE324 pKa = 3.76EE325 pKa = 5.29DD326 pKa = 3.88YY327 pKa = 11.58ALKK330 pKa = 10.56NYY332 pKa = 10.37KK333 pKa = 9.73LYY335 pKa = 10.84KK336 pKa = 9.06YY337 pKa = 10.08IRR339 pKa = 11.84RR340 pKa = 3.86
Molecular weight: 41.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1440 |
189 |
560 |
360.0 |
41.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 1.598 | 1.667 ± 0.451 |
6.25 ± 0.354 | 4.653 ± 1.205 |
5.903 ± 0.865 | 5.139 ± 0.86 |
2.014 ± 0.469 | 6.667 ± 0.35 |
7.083 ± 1.569 | 7.153 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.336 | 7.222 ± 0.899 |
4.375 ± 0.797 | 3.611 ± 0.308 |
3.958 ± 0.443 | 9.653 ± 1.662 |
4.653 ± 0.874 | 4.167 ± 0.67 |
0.833 ± 0.173 | 6.111 ± 0.626 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
