
Enterobacteria phage PRD1 (Bacteriophage PRD1)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Kalamavirales; Tectiviridae; Alphatectivirus; Pseudomonas virus PRD1
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
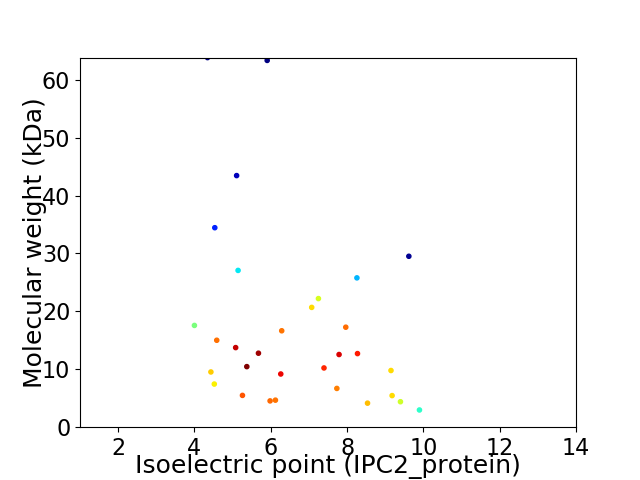
Virtual 2D-PAGE plot for 31 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P27379|PKG6_BPPRD Packaging efficiency factor P6 OS=Enterobacteria phage PRD1 OX=10658 GN=VI PE=1 SV=1
MM1 pKa = 7.78ANFNVPKK8 pKa = 10.64LGVFPVAAVFDD19 pKa = 4.02IDD21 pKa = 4.74NVPEE25 pKa = 4.55DD26 pKa = 3.71SSATGSRR33 pKa = 11.84WLPSIYY39 pKa = 10.04QGGNYY44 pKa = 8.86WGGGPQALHH53 pKa = 5.96AQVSNFDD60 pKa = 3.31SSNRR64 pKa = 11.84LPYY67 pKa = 10.4NPRR70 pKa = 11.84TEE72 pKa = 4.15NNPAGNCAFAFNPFGQYY89 pKa = 9.64ISNISSAQSVHH100 pKa = 5.09RR101 pKa = 11.84RR102 pKa = 11.84IYY104 pKa = 11.01GIDD107 pKa = 3.8LNDD110 pKa = 3.97EE111 pKa = 4.39PLFSPNAASITNGGNPTMSQDD132 pKa = 3.19TGYY135 pKa = 11.27HH136 pKa = 5.98NIGPINTAYY145 pKa = 9.79KK146 pKa = 10.74AEE148 pKa = 4.19IFRR151 pKa = 11.84PVNPLPMSDD160 pKa = 3.49TAPDD164 pKa = 4.21PEE166 pKa = 4.46TLEE169 pKa = 4.57PGQTEE174 pKa = 4.22PLIKK178 pKa = 10.24SDD180 pKa = 3.46GVYY183 pKa = 10.43SNSGIASFIFDD194 pKa = 3.96RR195 pKa = 11.84PVTEE199 pKa = 5.45PNPNWPPLPPPVIPIIYY216 pKa = 6.87PTPALGIGAAAAYY229 pKa = 9.77GFGYY233 pKa = 10.3QVTVYY238 pKa = 8.93RR239 pKa = 11.84WEE241 pKa = 4.45EE242 pKa = 3.81IPVEE246 pKa = 4.9FIADD250 pKa = 3.91PEE252 pKa = 4.38TCPAQPTTDD261 pKa = 3.08KK262 pKa = 11.31VIIRR266 pKa = 11.84TTDD269 pKa = 3.48LNPEE273 pKa = 4.52GSPCAYY279 pKa = 8.77EE280 pKa = 3.86AGIILVRR287 pKa = 11.84QTSNPMNAVAGRR299 pKa = 11.84LVPYY303 pKa = 10.64VEE305 pKa = 6.0DD306 pKa = 3.35IAVDD310 pKa = 3.56IFLTGKK316 pKa = 10.19FFTLNPPLRR325 pKa = 11.84ITNNYY330 pKa = 9.15FADD333 pKa = 4.21DD334 pKa = 3.89EE335 pKa = 4.81VKK337 pKa = 10.83EE338 pKa = 4.11NTVTIGNYY346 pKa = 6.41TTTLSSAYY354 pKa = 8.91YY355 pKa = 10.27AVYY358 pKa = 8.53KK359 pKa = 9.56TDD361 pKa = 3.53GYY363 pKa = 11.61GGATCFIASGGAGISALVQLQDD385 pKa = 3.44NSVLDD390 pKa = 3.49VLYY393 pKa = 11.0YY394 pKa = 10.57SLPLSLGGSKK404 pKa = 10.3AAIDD408 pKa = 3.16EE409 pKa = 4.44WVANNCGLFPMSGGLDD425 pKa = 3.11KK426 pKa = 9.33TTLLEE431 pKa = 3.84IPRR434 pKa = 11.84RR435 pKa = 11.84QLEE438 pKa = 4.58AINPQDD444 pKa = 4.42GPGQYY449 pKa = 11.06DD450 pKa = 4.27LFILDD455 pKa = 4.5DD456 pKa = 4.03SGAYY460 pKa = 10.15ASFSSFIGYY469 pKa = 8.9PEE471 pKa = 3.77AAYY474 pKa = 10.24YY475 pKa = 9.7VAGAATFMDD484 pKa = 4.13VEE486 pKa = 4.52NPDD489 pKa = 3.8EE490 pKa = 4.83IIFILRR496 pKa = 11.84NGAGWYY502 pKa = 10.04ACEE505 pKa = 4.86IGDD508 pKa = 4.32ALKK511 pKa = 10.62IADD514 pKa = 4.81DD515 pKa = 4.18EE516 pKa = 4.74FDD518 pKa = 3.65SVDD521 pKa = 3.22YY522 pKa = 9.99FAYY525 pKa = 10.27RR526 pKa = 11.84GGVMFIGSARR536 pKa = 11.84YY537 pKa = 7.72TEE539 pKa = 5.15GGDD542 pKa = 3.62PLPIKK547 pKa = 10.4YY548 pKa = 9.76RR549 pKa = 11.84AIIPGLPRR557 pKa = 11.84GRR559 pKa = 11.84LPRR562 pKa = 11.84VVLEE566 pKa = 3.96YY567 pKa = 10.53QAVGMSFIPCQTHH580 pKa = 6.33CLGKK584 pKa = 10.66GGIISKK590 pKa = 9.75VV591 pKa = 3.09
MM1 pKa = 7.78ANFNVPKK8 pKa = 10.64LGVFPVAAVFDD19 pKa = 4.02IDD21 pKa = 4.74NVPEE25 pKa = 4.55DD26 pKa = 3.71SSATGSRR33 pKa = 11.84WLPSIYY39 pKa = 10.04QGGNYY44 pKa = 8.86WGGGPQALHH53 pKa = 5.96AQVSNFDD60 pKa = 3.31SSNRR64 pKa = 11.84LPYY67 pKa = 10.4NPRR70 pKa = 11.84TEE72 pKa = 4.15NNPAGNCAFAFNPFGQYY89 pKa = 9.64ISNISSAQSVHH100 pKa = 5.09RR101 pKa = 11.84RR102 pKa = 11.84IYY104 pKa = 11.01GIDD107 pKa = 3.8LNDD110 pKa = 3.97EE111 pKa = 4.39PLFSPNAASITNGGNPTMSQDD132 pKa = 3.19TGYY135 pKa = 11.27HH136 pKa = 5.98NIGPINTAYY145 pKa = 9.79KK146 pKa = 10.74AEE148 pKa = 4.19IFRR151 pKa = 11.84PVNPLPMSDD160 pKa = 3.49TAPDD164 pKa = 4.21PEE166 pKa = 4.46TLEE169 pKa = 4.57PGQTEE174 pKa = 4.22PLIKK178 pKa = 10.24SDD180 pKa = 3.46GVYY183 pKa = 10.43SNSGIASFIFDD194 pKa = 3.96RR195 pKa = 11.84PVTEE199 pKa = 5.45PNPNWPPLPPPVIPIIYY216 pKa = 6.87PTPALGIGAAAAYY229 pKa = 9.77GFGYY233 pKa = 10.3QVTVYY238 pKa = 8.93RR239 pKa = 11.84WEE241 pKa = 4.45EE242 pKa = 3.81IPVEE246 pKa = 4.9FIADD250 pKa = 3.91PEE252 pKa = 4.38TCPAQPTTDD261 pKa = 3.08KK262 pKa = 11.31VIIRR266 pKa = 11.84TTDD269 pKa = 3.48LNPEE273 pKa = 4.52GSPCAYY279 pKa = 8.77EE280 pKa = 3.86AGIILVRR287 pKa = 11.84QTSNPMNAVAGRR299 pKa = 11.84LVPYY303 pKa = 10.64VEE305 pKa = 6.0DD306 pKa = 3.35IAVDD310 pKa = 3.56IFLTGKK316 pKa = 10.19FFTLNPPLRR325 pKa = 11.84ITNNYY330 pKa = 9.15FADD333 pKa = 4.21DD334 pKa = 3.89EE335 pKa = 4.81VKK337 pKa = 10.83EE338 pKa = 4.11NTVTIGNYY346 pKa = 6.41TTTLSSAYY354 pKa = 8.91YY355 pKa = 10.27AVYY358 pKa = 8.53KK359 pKa = 9.56TDD361 pKa = 3.53GYY363 pKa = 11.61GGATCFIASGGAGISALVQLQDD385 pKa = 3.44NSVLDD390 pKa = 3.49VLYY393 pKa = 11.0YY394 pKa = 10.57SLPLSLGGSKK404 pKa = 10.3AAIDD408 pKa = 3.16EE409 pKa = 4.44WVANNCGLFPMSGGLDD425 pKa = 3.11KK426 pKa = 9.33TTLLEE431 pKa = 3.84IPRR434 pKa = 11.84RR435 pKa = 11.84QLEE438 pKa = 4.58AINPQDD444 pKa = 4.42GPGQYY449 pKa = 11.06DD450 pKa = 4.27LFILDD455 pKa = 4.5DD456 pKa = 4.03SGAYY460 pKa = 10.15ASFSSFIGYY469 pKa = 8.9PEE471 pKa = 3.77AAYY474 pKa = 10.24YY475 pKa = 9.7VAGAATFMDD484 pKa = 4.13VEE486 pKa = 4.52NPDD489 pKa = 3.8EE490 pKa = 4.83IIFILRR496 pKa = 11.84NGAGWYY502 pKa = 10.04ACEE505 pKa = 4.86IGDD508 pKa = 4.32ALKK511 pKa = 10.62IADD514 pKa = 4.81DD515 pKa = 4.18EE516 pKa = 4.74FDD518 pKa = 3.65SVDD521 pKa = 3.22YY522 pKa = 9.99FAYY525 pKa = 10.27RR526 pKa = 11.84GGVMFIGSARR536 pKa = 11.84YY537 pKa = 7.72TEE539 pKa = 5.15GGDD542 pKa = 3.62PLPIKK547 pKa = 10.4YY548 pKa = 9.76RR549 pKa = 11.84AIIPGLPRR557 pKa = 11.84GRR559 pKa = 11.84LPRR562 pKa = 11.84VVLEE566 pKa = 3.96YY567 pKa = 10.53QAVGMSFIPCQTHH580 pKa = 6.33CLGKK584 pKa = 10.66GGIISKK590 pKa = 9.75VV591 pKa = 3.09
Molecular weight: 63.82 kDa
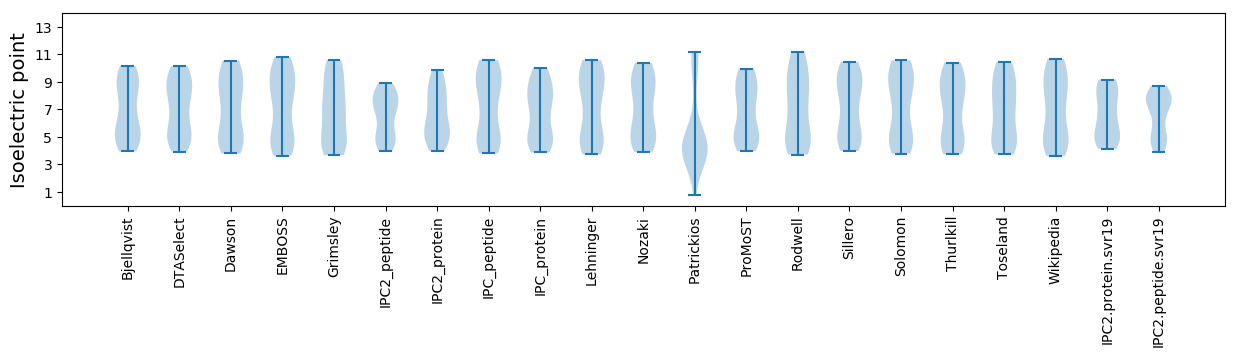
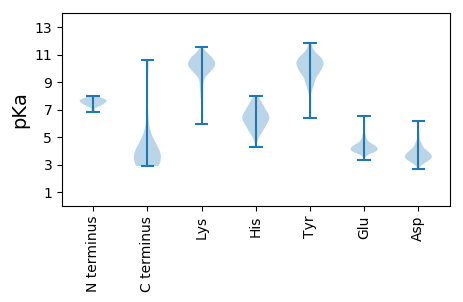
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q3T4N8|VP33_BPPRD Protein P33 OS=Enterobacteria phage PRD1 OX=10658 GN=XXXIII PE=4 SV=1
MM1 pKa = 7.77KK2 pKa = 10.22IFGISVFTIAIIALAYY18 pKa = 10.19YY19 pKa = 10.3LGTKK23 pKa = 10.15RR24 pKa = 11.84IIGG27 pKa = 3.68
MM1 pKa = 7.77KK2 pKa = 10.22IFGISVFTIAIIALAYY18 pKa = 10.19YY19 pKa = 10.3LGTKK23 pKa = 10.15RR24 pKa = 11.84IIGG27 pKa = 3.68
Molecular weight: 2.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5013 |
27 |
591 |
161.7 |
17.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.55 ± 0.893 | 0.798 ± 0.18 |
4.668 ± 0.416 | 5.246 ± 0.663 |
4.169 ± 0.438 | 8.777 ± 0.614 |
0.977 ± 0.233 | 6.782 ± 0.49 |
5.306 ± 0.711 | 7.8 ± 0.438 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.733 ± 0.278 | 5.067 ± 0.363 |
5.127 ± 0.541 | 4.169 ± 0.611 |
3.75 ± 0.351 | 5.645 ± 0.467 |
5.665 ± 0.426 | 6.723 ± 0.39 |
1.237 ± 0.144 | 3.81 ± 0.378 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
