
Cellvibrio sp. PSBB006
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Cellvibrio; unclassified Cellvibrio
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
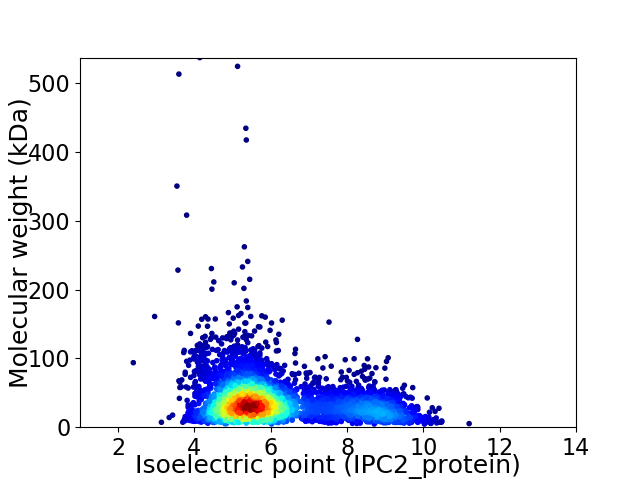
Virtual 2D-PAGE plot for 4268 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0FSD2|A0A1Y0FSD2_9GAMM Agmatine deiminase OS=Cellvibrio sp. PSBB006 OX=1987723 GN=CBR65_01995 PE=4 SV=1
MM1 pKa = 7.25FKK3 pKa = 10.54VLPILLSVFLLISCGGGGGGGSSDD27 pKa = 3.35GGEE30 pKa = 4.35TRR32 pKa = 11.84SSTSSSSSSSEE43 pKa = 3.78SSSSVTSSSQSSSSDD58 pKa = 3.02SSSSVAPNEE67 pKa = 4.15APTASAGEE75 pKa = 4.01DD76 pKa = 3.28QIVQGGFTVQLTAAGSADD94 pKa = 3.72PEE96 pKa = 4.89GEE98 pKa = 4.1TLTYY102 pKa = 10.61EE103 pKa = 4.18WMQVTEE109 pKa = 4.21HH110 pKa = 6.69NIMYY114 pKa = 8.01VTADD118 pKa = 3.4GTITEE123 pKa = 4.48EE124 pKa = 4.81MDD126 pKa = 3.41TQDD129 pKa = 3.59LFVYY133 pKa = 10.46IPATEE138 pKa = 4.05AGKK141 pKa = 10.59EE142 pKa = 4.08YY143 pKa = 10.14TFSLTVSDD151 pKa = 4.12GTHH154 pKa = 5.54TSEE157 pKa = 5.21ADD159 pKa = 3.51TVTLTVADD167 pKa = 4.3CFEE170 pKa = 5.17GDD172 pKa = 2.81GDD174 pKa = 4.75VFIEE178 pKa = 5.32CIDD181 pKa = 3.92PSWFGISSYY190 pKa = 9.88QEE192 pKa = 3.39IDD194 pKa = 3.23GNAGNNFYY202 pKa = 11.38SNGIDD207 pKa = 3.31NHH209 pKa = 6.3VNWSLVDD216 pKa = 4.1LEE218 pKa = 5.36DD219 pKa = 4.69GEE221 pKa = 4.85HH222 pKa = 6.55NNVIDD227 pKa = 4.7VKK229 pKa = 10.2FLRR232 pKa = 11.84NDD234 pKa = 2.8SSGNFTIAAPTPEE247 pKa = 4.47NADD250 pKa = 3.35MPAIDD255 pKa = 4.01MTDD258 pKa = 3.94FANGYY263 pKa = 9.73IKK265 pKa = 10.57FDD267 pKa = 3.41VRR269 pKa = 11.84AVGTTDD275 pKa = 3.62PLTLFLSIQCGYY287 pKa = 9.66PCGTSTPWTVEE298 pKa = 3.95TLPDD302 pKa = 3.99SEE304 pKa = 4.4WTSVTIWVEE313 pKa = 3.68NLIDD317 pKa = 4.86LEE319 pKa = 4.56LDD321 pKa = 3.3VSKK324 pKa = 10.91ISTALLINPQWVNQADD340 pKa = 3.9VNFQLDD346 pKa = 3.64NIRR349 pKa = 11.84WEE351 pKa = 4.24KK352 pKa = 10.78ASCDD356 pKa = 3.58DD357 pKa = 3.71APNVVFSNCLIQDD370 pKa = 3.75DD371 pKa = 4.71RR372 pKa = 11.84SASGWDD378 pKa = 3.44EE379 pKa = 4.88DD380 pKa = 3.81GGFYY384 pKa = 10.92GLPDD388 pKa = 4.87PDD390 pKa = 4.26NHH392 pKa = 6.64ISWLEE397 pKa = 3.79IFSGDD402 pKa = 3.5EE403 pKa = 3.67GHH405 pKa = 6.68NDD407 pKa = 3.32VIEE410 pKa = 4.03VSFNNDD416 pKa = 2.9TQSSSFSIIPVPLASMDD433 pKa = 3.91LSAYY437 pKa = 10.07AAGEE441 pKa = 4.05MVFDD445 pKa = 4.36YY446 pKa = 11.47KK447 pKa = 10.55MVSNPHH453 pKa = 6.8ADD455 pKa = 3.38TNLFATVQCGDD466 pKa = 3.51WPCAGGEE473 pKa = 3.99FLLDD477 pKa = 3.37KK478 pKa = 11.33SEE480 pKa = 4.29FGVWKK485 pKa = 10.19EE486 pKa = 3.82MRR488 pKa = 11.84ISVADD493 pKa = 4.17MIDD496 pKa = 3.25NDD498 pKa = 4.3LDD500 pKa = 3.49IMQVSTWFTLIPTYY514 pKa = 10.23GQPQSGIRR522 pKa = 11.84VEE524 pKa = 4.05LDD526 pKa = 3.12NIRR529 pKa = 11.84WEE531 pKa = 4.21EE532 pKa = 3.78
MM1 pKa = 7.25FKK3 pKa = 10.54VLPILLSVFLLISCGGGGGGGSSDD27 pKa = 3.35GGEE30 pKa = 4.35TRR32 pKa = 11.84SSTSSSSSSSEE43 pKa = 3.78SSSSVTSSSQSSSSDD58 pKa = 3.02SSSSVAPNEE67 pKa = 4.15APTASAGEE75 pKa = 4.01DD76 pKa = 3.28QIVQGGFTVQLTAAGSADD94 pKa = 3.72PEE96 pKa = 4.89GEE98 pKa = 4.1TLTYY102 pKa = 10.61EE103 pKa = 4.18WMQVTEE109 pKa = 4.21HH110 pKa = 6.69NIMYY114 pKa = 8.01VTADD118 pKa = 3.4GTITEE123 pKa = 4.48EE124 pKa = 4.81MDD126 pKa = 3.41TQDD129 pKa = 3.59LFVYY133 pKa = 10.46IPATEE138 pKa = 4.05AGKK141 pKa = 10.59EE142 pKa = 4.08YY143 pKa = 10.14TFSLTVSDD151 pKa = 4.12GTHH154 pKa = 5.54TSEE157 pKa = 5.21ADD159 pKa = 3.51TVTLTVADD167 pKa = 4.3CFEE170 pKa = 5.17GDD172 pKa = 2.81GDD174 pKa = 4.75VFIEE178 pKa = 5.32CIDD181 pKa = 3.92PSWFGISSYY190 pKa = 9.88QEE192 pKa = 3.39IDD194 pKa = 3.23GNAGNNFYY202 pKa = 11.38SNGIDD207 pKa = 3.31NHH209 pKa = 6.3VNWSLVDD216 pKa = 4.1LEE218 pKa = 5.36DD219 pKa = 4.69GEE221 pKa = 4.85HH222 pKa = 6.55NNVIDD227 pKa = 4.7VKK229 pKa = 10.2FLRR232 pKa = 11.84NDD234 pKa = 2.8SSGNFTIAAPTPEE247 pKa = 4.47NADD250 pKa = 3.35MPAIDD255 pKa = 4.01MTDD258 pKa = 3.94FANGYY263 pKa = 9.73IKK265 pKa = 10.57FDD267 pKa = 3.41VRR269 pKa = 11.84AVGTTDD275 pKa = 3.62PLTLFLSIQCGYY287 pKa = 9.66PCGTSTPWTVEE298 pKa = 3.95TLPDD302 pKa = 3.99SEE304 pKa = 4.4WTSVTIWVEE313 pKa = 3.68NLIDD317 pKa = 4.86LEE319 pKa = 4.56LDD321 pKa = 3.3VSKK324 pKa = 10.91ISTALLINPQWVNQADD340 pKa = 3.9VNFQLDD346 pKa = 3.64NIRR349 pKa = 11.84WEE351 pKa = 4.24KK352 pKa = 10.78ASCDD356 pKa = 3.58DD357 pKa = 3.71APNVVFSNCLIQDD370 pKa = 3.75DD371 pKa = 4.71RR372 pKa = 11.84SASGWDD378 pKa = 3.44EE379 pKa = 4.88DD380 pKa = 3.81GGFYY384 pKa = 10.92GLPDD388 pKa = 4.87PDD390 pKa = 4.26NHH392 pKa = 6.64ISWLEE397 pKa = 3.79IFSGDD402 pKa = 3.5EE403 pKa = 3.67GHH405 pKa = 6.68NDD407 pKa = 3.32VIEE410 pKa = 4.03VSFNNDD416 pKa = 2.9TQSSSFSIIPVPLASMDD433 pKa = 3.91LSAYY437 pKa = 10.07AAGEE441 pKa = 4.05MVFDD445 pKa = 4.36YY446 pKa = 11.47KK447 pKa = 10.55MVSNPHH453 pKa = 6.8ADD455 pKa = 3.38TNLFATVQCGDD466 pKa = 3.51WPCAGGEE473 pKa = 3.99FLLDD477 pKa = 3.37KK478 pKa = 11.33SEE480 pKa = 4.29FGVWKK485 pKa = 10.19EE486 pKa = 3.82MRR488 pKa = 11.84ISVADD493 pKa = 4.17MIDD496 pKa = 3.25NDD498 pKa = 4.3LDD500 pKa = 3.49IMQVSTWFTLIPTYY514 pKa = 10.23GQPQSGIRR522 pKa = 11.84VEE524 pKa = 4.05LDD526 pKa = 3.12NIRR529 pKa = 11.84WEE531 pKa = 4.21EE532 pKa = 3.78
Molecular weight: 57.66 kDa
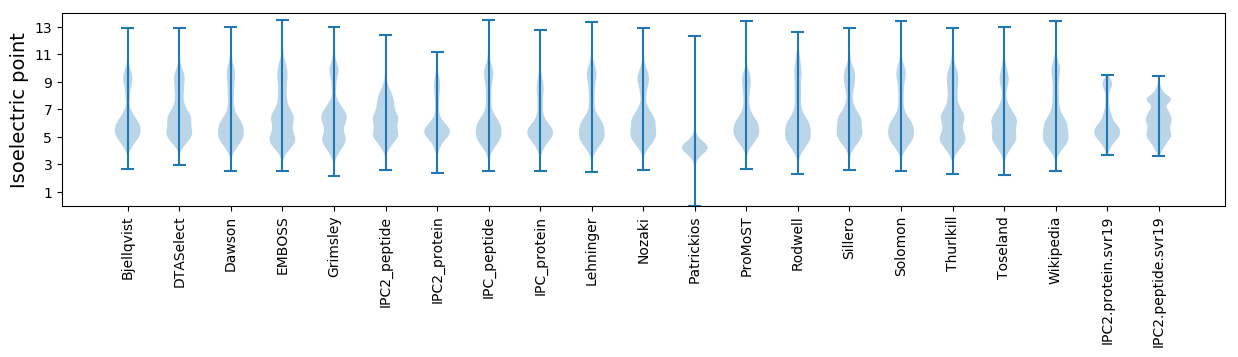
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0FYY1|A0A1Y0FYY1_9GAMM Uncharacterized protein OS=Cellvibrio sp. PSBB006 OX=1987723 GN=CBR65_15130 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.95RR12 pKa = 11.84VRR14 pKa = 11.84NHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 8.34QLTII44 pKa = 3.64
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.95RR12 pKa = 11.84VRR14 pKa = 11.84NHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 8.34QLTII44 pKa = 3.64
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1542520 |
38 |
5127 |
361.4 |
40.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.426 ± 0.041 | 0.935 ± 0.013 |
5.799 ± 0.035 | 5.878 ± 0.034 |
3.981 ± 0.024 | 7.174 ± 0.054 |
2.304 ± 0.024 | 5.898 ± 0.027 |
4.098 ± 0.039 | 10.345 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.018 | 4.242 ± 0.05 |
4.376 ± 0.025 | 4.445 ± 0.033 |
5.397 ± 0.038 | 6.396 ± 0.042 |
5.538 ± 0.04 | 6.895 ± 0.028 |
1.465 ± 0.018 | 3.099 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
