
Chondrostereum purpureum cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
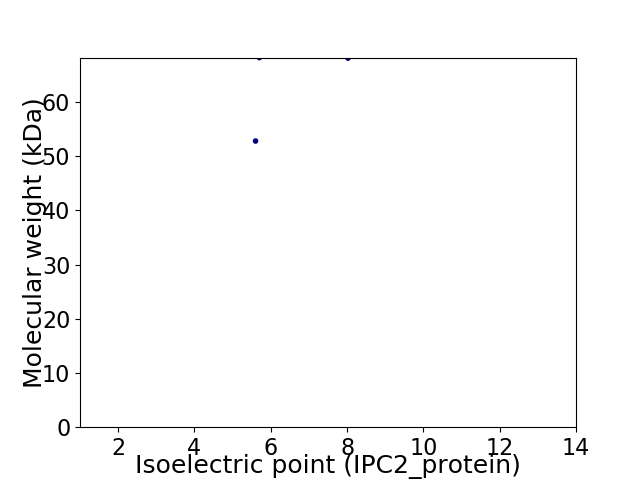
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B7ZGY9|B7ZGY9_9VIRU Coat protein OS=Chondrostereum purpureum cryptic virus 1 OX=529380 GN=CP PE=4 SV=1
MM1 pKa = 7.34SLSSGPVSQAVLTASEE17 pKa = 4.38PAVPAPVEE25 pKa = 4.06YY26 pKa = 10.36RR27 pKa = 11.84PGATQPDD34 pKa = 3.79ATGTMTQTASGTANAIFQSGATTAPEE60 pKa = 3.83QPPVLIHH67 pKa = 6.52VSDD70 pKa = 4.08DD71 pKa = 3.41AKK73 pKa = 11.44SNEE76 pKa = 4.06KK77 pKa = 9.91MSAIDD82 pKa = 3.05ITRR85 pKa = 11.84IAICYY90 pKa = 8.87PMSTDD95 pKa = 3.48SVLVPSWFLPSSRR108 pKa = 11.84YY109 pKa = 9.57LYY111 pKa = 10.4RR112 pKa = 11.84IVQEE116 pKa = 3.91MDD118 pKa = 2.87IIMLNTHH125 pKa = 7.15RR126 pKa = 11.84FTQSVHH132 pKa = 4.79AWSPLVSRR140 pKa = 11.84VYY142 pKa = 10.2IAVLFWIQTIRR153 pKa = 11.84CMQATGFLPRR163 pKa = 11.84EE164 pKa = 4.46GIAFINEE171 pKa = 4.22LLQDD175 pKa = 4.3FPARR179 pKa = 11.84TLVVPGPLVSLFRR192 pKa = 11.84SLCASSPSFGTYY204 pKa = 10.45GDD206 pKa = 3.7VFPAFEE212 pKa = 4.12TAGIGASPANCFTLNSARR230 pKa = 11.84FPQWYY235 pKa = 8.37MLMPNVALLLDD246 pKa = 3.64QLTCFTQACVNNPATIGDD264 pKa = 4.02FVIGDD269 pKa = 4.34DD270 pKa = 4.91LFGTPFDD277 pKa = 3.93GTIGSHH283 pKa = 5.01RR284 pKa = 11.84QQRR287 pKa = 11.84VAPGMTEE294 pKa = 3.4PVYY297 pKa = 10.86FNTKK301 pKa = 9.35AAEE304 pKa = 3.98NHH306 pKa = 5.36ASMRR310 pKa = 11.84NLMALPTRR318 pKa = 11.84LNYY321 pKa = 10.38DD322 pKa = 3.53PQNATPLTLNEE333 pKa = 4.02MFRR336 pKa = 11.84FNTDD340 pKa = 2.41KK341 pKa = 10.85RR342 pKa = 11.84WFSRR346 pKa = 11.84IISTMTVYY354 pKa = 10.95SKK356 pKa = 10.97FFDD359 pKa = 4.15GSVTLYY365 pKa = 9.12EE366 pKa = 4.82CPPVGPPAAQVSMVRR381 pKa = 11.84TQPMRR386 pKa = 11.84FEE388 pKa = 4.76ASNTAQAMEE397 pKa = 4.38TEE399 pKa = 4.69SFEE402 pKa = 4.21STAISVNRR410 pKa = 11.84SLSLHH415 pKa = 6.02DD416 pKa = 4.26EE417 pKa = 4.32KK418 pKa = 11.52VALAALINEE427 pKa = 4.61TLVSTGDD434 pKa = 3.46QPIYY438 pKa = 10.06AAQRR442 pKa = 11.84LGPFWNISPTRR453 pKa = 11.84RR454 pKa = 11.84QAYY457 pKa = 9.4RR458 pKa = 11.84FDD460 pKa = 3.87PSFVLPSVIRR470 pKa = 11.84DD471 pKa = 3.75HH472 pKa = 6.79YY473 pKa = 11.71ALTTATRR480 pKa = 3.54
MM1 pKa = 7.34SLSSGPVSQAVLTASEE17 pKa = 4.38PAVPAPVEE25 pKa = 4.06YY26 pKa = 10.36RR27 pKa = 11.84PGATQPDD34 pKa = 3.79ATGTMTQTASGTANAIFQSGATTAPEE60 pKa = 3.83QPPVLIHH67 pKa = 6.52VSDD70 pKa = 4.08DD71 pKa = 3.41AKK73 pKa = 11.44SNEE76 pKa = 4.06KK77 pKa = 9.91MSAIDD82 pKa = 3.05ITRR85 pKa = 11.84IAICYY90 pKa = 8.87PMSTDD95 pKa = 3.48SVLVPSWFLPSSRR108 pKa = 11.84YY109 pKa = 9.57LYY111 pKa = 10.4RR112 pKa = 11.84IVQEE116 pKa = 3.91MDD118 pKa = 2.87IIMLNTHH125 pKa = 7.15RR126 pKa = 11.84FTQSVHH132 pKa = 4.79AWSPLVSRR140 pKa = 11.84VYY142 pKa = 10.2IAVLFWIQTIRR153 pKa = 11.84CMQATGFLPRR163 pKa = 11.84EE164 pKa = 4.46GIAFINEE171 pKa = 4.22LLQDD175 pKa = 4.3FPARR179 pKa = 11.84TLVVPGPLVSLFRR192 pKa = 11.84SLCASSPSFGTYY204 pKa = 10.45GDD206 pKa = 3.7VFPAFEE212 pKa = 4.12TAGIGASPANCFTLNSARR230 pKa = 11.84FPQWYY235 pKa = 8.37MLMPNVALLLDD246 pKa = 3.64QLTCFTQACVNNPATIGDD264 pKa = 4.02FVIGDD269 pKa = 4.34DD270 pKa = 4.91LFGTPFDD277 pKa = 3.93GTIGSHH283 pKa = 5.01RR284 pKa = 11.84QQRR287 pKa = 11.84VAPGMTEE294 pKa = 3.4PVYY297 pKa = 10.86FNTKK301 pKa = 9.35AAEE304 pKa = 3.98NHH306 pKa = 5.36ASMRR310 pKa = 11.84NLMALPTRR318 pKa = 11.84LNYY321 pKa = 10.38DD322 pKa = 3.53PQNATPLTLNEE333 pKa = 4.02MFRR336 pKa = 11.84FNTDD340 pKa = 2.41KK341 pKa = 10.85RR342 pKa = 11.84WFSRR346 pKa = 11.84IISTMTVYY354 pKa = 10.95SKK356 pKa = 10.97FFDD359 pKa = 4.15GSVTLYY365 pKa = 9.12EE366 pKa = 4.82CPPVGPPAAQVSMVRR381 pKa = 11.84TQPMRR386 pKa = 11.84FEE388 pKa = 4.76ASNTAQAMEE397 pKa = 4.38TEE399 pKa = 4.69SFEE402 pKa = 4.21STAISVNRR410 pKa = 11.84SLSLHH415 pKa = 6.02DD416 pKa = 4.26EE417 pKa = 4.32KK418 pKa = 11.52VALAALINEE427 pKa = 4.61TLVSTGDD434 pKa = 3.46QPIYY438 pKa = 10.06AAQRR442 pKa = 11.84LGPFWNISPTRR453 pKa = 11.84RR454 pKa = 11.84QAYY457 pKa = 9.4RR458 pKa = 11.84FDD460 pKa = 3.87PSFVLPSVIRR470 pKa = 11.84DD471 pKa = 3.75HH472 pKa = 6.79YY473 pKa = 11.71ALTTATRR480 pKa = 3.54
Molecular weight: 52.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B7ZGY9|B7ZGY9_9VIRU Coat protein OS=Chondrostereum purpureum cryptic virus 1 OX=529380 GN=CP PE=4 SV=1
MM1 pKa = 7.95RR2 pKa = 11.84FTEE5 pKa = 4.44YY6 pKa = 10.19ILSGFKK12 pKa = 10.84VLDD15 pKa = 3.48SSMDD19 pKa = 3.13RR20 pKa = 11.84SRR22 pKa = 11.84NYY24 pKa = 10.46QFIGFDD30 pKa = 3.61KK31 pKa = 11.0PEE33 pKa = 4.25SQPPADD39 pKa = 3.71ATMHH43 pKa = 6.5HH44 pKa = 7.03AVQAPVKK51 pKa = 9.59HH52 pKa = 6.15AMRR55 pKa = 11.84KK56 pKa = 8.85ILLHH60 pKa = 6.22NEE62 pKa = 4.01FVHH65 pKa = 5.8ITTKK69 pKa = 10.66FKK71 pKa = 10.87RR72 pKa = 11.84STIDD76 pKa = 3.22DD77 pKa = 3.74SSITEE82 pKa = 4.15AFFKK86 pKa = 11.09GDD88 pKa = 3.57LPYY91 pKa = 10.99HH92 pKa = 6.38KK93 pKa = 10.26VPKK96 pKa = 10.19DD97 pKa = 3.36GSYY100 pKa = 10.8QRR102 pKa = 11.84ALAATMQVFAPQVPIKK118 pKa = 9.72PVHH121 pKa = 5.6YY122 pKa = 9.95CDD124 pKa = 4.32LRR126 pKa = 11.84YY127 pKa = 10.18YY128 pKa = 9.15PWKK131 pKa = 10.17LRR133 pKa = 11.84PSAEE137 pKa = 4.58LPFTDD142 pKa = 5.7DD143 pKa = 3.13PALFKK148 pKa = 10.5RR149 pKa = 11.84VQHH152 pKa = 5.54AAEE155 pKa = 4.51RR156 pKa = 11.84KK157 pKa = 8.91QIPDD161 pKa = 2.79KK162 pKa = 11.15RR163 pKa = 11.84MSFGNLYY170 pKa = 10.35NHH172 pKa = 6.65IFEE175 pKa = 4.3YY176 pKa = 10.42LRR178 pKa = 11.84PIIHH182 pKa = 6.55YY183 pKa = 9.38IKK185 pKa = 10.57NGPFHH190 pKa = 7.03LPFKK194 pKa = 10.02TSWPQDD200 pKa = 3.45DD201 pKa = 5.14YY202 pKa = 10.25LTPMRR207 pKa = 11.84SHH209 pKa = 6.71ARR211 pKa = 11.84SAIQPISDD219 pKa = 3.73PNKK222 pKa = 7.97VRR224 pKa = 11.84FVYY227 pKa = 9.7GYY229 pKa = 9.01PKK231 pKa = 10.72VGIFPQAMFFWPLFVYY247 pKa = 8.24YY248 pKa = 9.9HH249 pKa = 6.32ITGQSPLLWGYY260 pKa = 7.75EE261 pKa = 3.99TMTGGWYY268 pKa = 10.39RR269 pKa = 11.84LNHH272 pKa = 6.05EE273 pKa = 4.67LRR275 pKa = 11.84STPGISGSILEE286 pKa = 4.97LDD288 pKa = 3.29WQGFDD293 pKa = 2.56IHH295 pKa = 8.38ALFEE299 pKa = 4.3VFDD302 pKa = 5.14DD303 pKa = 4.17IKK305 pKa = 11.04NLSWSFFDD313 pKa = 5.15CSHH316 pKa = 6.9GYY318 pKa = 9.79QPTKK322 pKa = 10.53DD323 pKa = 3.89YY324 pKa = 11.37QGTSCDD330 pKa = 3.57PDD332 pKa = 3.21RR333 pKa = 11.84LKK335 pKa = 11.14NLWEE339 pKa = 4.6WIWFSIKK346 pKa = 9.1YY347 pKa = 8.22TPIRR351 pKa = 11.84LPDD354 pKa = 3.13GRR356 pKa = 11.84LFRR359 pKa = 11.84RR360 pKa = 11.84SWRR363 pKa = 11.84GVPSGLFITQWLDD376 pKa = 3.24SVYY379 pKa = 9.86NCIMIFTILDD389 pKa = 3.24AMGFEE394 pKa = 4.4ITSDD398 pKa = 4.34LIIKK402 pKa = 9.68VLGDD406 pKa = 3.79DD407 pKa = 3.33SLTRR411 pKa = 11.84LRR413 pKa = 11.84LLIPYY418 pKa = 7.94DD419 pKa = 3.17QHH421 pKa = 5.73EE422 pKa = 4.5AFTIRR427 pKa = 11.84FKK429 pKa = 11.15HH430 pKa = 5.21LAQYY434 pKa = 10.95YY435 pKa = 9.21FDD437 pKa = 5.39AVLSDD442 pKa = 3.39KK443 pKa = 10.66KK444 pKa = 11.22SRR446 pKa = 11.84IHH448 pKa = 5.58NTLNGVKK455 pKa = 10.42CLGYY459 pKa = 10.39RR460 pKa = 11.84NNNGLPVRR468 pKa = 11.84DD469 pKa = 3.92RR470 pKa = 11.84EE471 pKa = 4.41SLLTALLYY479 pKa = 10.31PKK481 pKa = 10.28ARR483 pKa = 11.84RR484 pKa = 11.84PTFEE488 pKa = 3.85QLKK491 pKa = 9.86ARR493 pKa = 11.84CIGIAYY499 pKa = 9.73ASAGNDD505 pKa = 3.21PTTLEE510 pKa = 4.08VCKK513 pKa = 10.53DD514 pKa = 3.26VYY516 pKa = 11.23DD517 pKa = 3.95YY518 pKa = 11.88LDD520 pKa = 3.63AKK522 pKa = 10.2GIKK525 pKa = 9.72ADD527 pKa = 3.87PAGLADD533 pKa = 5.77LFDD536 pKa = 5.73PNLGLDD542 pKa = 3.11IDD544 pKa = 3.79ITFFPNILTIQRR556 pKa = 11.84SLLILRR562 pKa = 11.84DD563 pKa = 3.52SRR565 pKa = 11.84EE566 pKa = 3.81QAHH569 pKa = 6.4RR570 pKa = 11.84MWPTSHH576 pKa = 7.23FLSPIPGLVDD586 pKa = 3.12II587 pKa = 5.55
MM1 pKa = 7.95RR2 pKa = 11.84FTEE5 pKa = 4.44YY6 pKa = 10.19ILSGFKK12 pKa = 10.84VLDD15 pKa = 3.48SSMDD19 pKa = 3.13RR20 pKa = 11.84SRR22 pKa = 11.84NYY24 pKa = 10.46QFIGFDD30 pKa = 3.61KK31 pKa = 11.0PEE33 pKa = 4.25SQPPADD39 pKa = 3.71ATMHH43 pKa = 6.5HH44 pKa = 7.03AVQAPVKK51 pKa = 9.59HH52 pKa = 6.15AMRR55 pKa = 11.84KK56 pKa = 8.85ILLHH60 pKa = 6.22NEE62 pKa = 4.01FVHH65 pKa = 5.8ITTKK69 pKa = 10.66FKK71 pKa = 10.87RR72 pKa = 11.84STIDD76 pKa = 3.22DD77 pKa = 3.74SSITEE82 pKa = 4.15AFFKK86 pKa = 11.09GDD88 pKa = 3.57LPYY91 pKa = 10.99HH92 pKa = 6.38KK93 pKa = 10.26VPKK96 pKa = 10.19DD97 pKa = 3.36GSYY100 pKa = 10.8QRR102 pKa = 11.84ALAATMQVFAPQVPIKK118 pKa = 9.72PVHH121 pKa = 5.6YY122 pKa = 9.95CDD124 pKa = 4.32LRR126 pKa = 11.84YY127 pKa = 10.18YY128 pKa = 9.15PWKK131 pKa = 10.17LRR133 pKa = 11.84PSAEE137 pKa = 4.58LPFTDD142 pKa = 5.7DD143 pKa = 3.13PALFKK148 pKa = 10.5RR149 pKa = 11.84VQHH152 pKa = 5.54AAEE155 pKa = 4.51RR156 pKa = 11.84KK157 pKa = 8.91QIPDD161 pKa = 2.79KK162 pKa = 11.15RR163 pKa = 11.84MSFGNLYY170 pKa = 10.35NHH172 pKa = 6.65IFEE175 pKa = 4.3YY176 pKa = 10.42LRR178 pKa = 11.84PIIHH182 pKa = 6.55YY183 pKa = 9.38IKK185 pKa = 10.57NGPFHH190 pKa = 7.03LPFKK194 pKa = 10.02TSWPQDD200 pKa = 3.45DD201 pKa = 5.14YY202 pKa = 10.25LTPMRR207 pKa = 11.84SHH209 pKa = 6.71ARR211 pKa = 11.84SAIQPISDD219 pKa = 3.73PNKK222 pKa = 7.97VRR224 pKa = 11.84FVYY227 pKa = 9.7GYY229 pKa = 9.01PKK231 pKa = 10.72VGIFPQAMFFWPLFVYY247 pKa = 8.24YY248 pKa = 9.9HH249 pKa = 6.32ITGQSPLLWGYY260 pKa = 7.75EE261 pKa = 3.99TMTGGWYY268 pKa = 10.39RR269 pKa = 11.84LNHH272 pKa = 6.05EE273 pKa = 4.67LRR275 pKa = 11.84STPGISGSILEE286 pKa = 4.97LDD288 pKa = 3.29WQGFDD293 pKa = 2.56IHH295 pKa = 8.38ALFEE299 pKa = 4.3VFDD302 pKa = 5.14DD303 pKa = 4.17IKK305 pKa = 11.04NLSWSFFDD313 pKa = 5.15CSHH316 pKa = 6.9GYY318 pKa = 9.79QPTKK322 pKa = 10.53DD323 pKa = 3.89YY324 pKa = 11.37QGTSCDD330 pKa = 3.57PDD332 pKa = 3.21RR333 pKa = 11.84LKK335 pKa = 11.14NLWEE339 pKa = 4.6WIWFSIKK346 pKa = 9.1YY347 pKa = 8.22TPIRR351 pKa = 11.84LPDD354 pKa = 3.13GRR356 pKa = 11.84LFRR359 pKa = 11.84RR360 pKa = 11.84SWRR363 pKa = 11.84GVPSGLFITQWLDD376 pKa = 3.24SVYY379 pKa = 9.86NCIMIFTILDD389 pKa = 3.24AMGFEE394 pKa = 4.4ITSDD398 pKa = 4.34LIIKK402 pKa = 9.68VLGDD406 pKa = 3.79DD407 pKa = 3.33SLTRR411 pKa = 11.84LRR413 pKa = 11.84LLIPYY418 pKa = 7.94DD419 pKa = 3.17QHH421 pKa = 5.73EE422 pKa = 4.5AFTIRR427 pKa = 11.84FKK429 pKa = 11.15HH430 pKa = 5.21LAQYY434 pKa = 10.95YY435 pKa = 9.21FDD437 pKa = 5.39AVLSDD442 pKa = 3.39KK443 pKa = 10.66KK444 pKa = 11.22SRR446 pKa = 11.84IHH448 pKa = 5.58NTLNGVKK455 pKa = 10.42CLGYY459 pKa = 10.39RR460 pKa = 11.84NNNGLPVRR468 pKa = 11.84DD469 pKa = 3.92RR470 pKa = 11.84EE471 pKa = 4.41SLLTALLYY479 pKa = 10.31PKK481 pKa = 10.28ARR483 pKa = 11.84RR484 pKa = 11.84PTFEE488 pKa = 3.85QLKK491 pKa = 9.86ARR493 pKa = 11.84CIGIAYY499 pKa = 9.73ASAGNDD505 pKa = 3.21PTTLEE510 pKa = 4.08VCKK513 pKa = 10.53DD514 pKa = 3.26VYY516 pKa = 11.23DD517 pKa = 3.95YY518 pKa = 11.88LDD520 pKa = 3.63AKK522 pKa = 10.2GIKK525 pKa = 9.72ADD527 pKa = 3.87PAGLADD533 pKa = 5.77LFDD536 pKa = 5.73PNLGLDD542 pKa = 3.11IDD544 pKa = 3.79ITFFPNILTIQRR556 pKa = 11.84SLLILRR562 pKa = 11.84DD563 pKa = 3.52SRR565 pKa = 11.84EE566 pKa = 3.81QAHH569 pKa = 6.4RR570 pKa = 11.84MWPTSHH576 pKa = 7.23FLSPIPGLVDD586 pKa = 3.12II587 pKa = 5.55
Molecular weight: 68.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1067 |
480 |
587 |
533.5 |
60.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.591 ± 1.53 | 1.312 ± 0.093 |
5.998 ± 1.163 | 3.374 ± 0.239 |
6.186 ± 0.224 | 4.967 ± 0.244 |
2.624 ± 0.741 | 6.373 ± 0.74 |
3.561 ± 1.468 | 8.903 ± 0.759 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.718 ± 0.523 | 3.561 ± 0.385 |
7.591 ± 0.339 | 4.03 ± 0.352 |
5.998 ± 0.237 | 7.404 ± 0.855 |
6.842 ± 1.345 | 5.155 ± 0.961 |
1.781 ± 0.337 | 4.03 ± 0.707 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
