
Sumatran orang-utan polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Pongo abelii polyomavirus 1
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
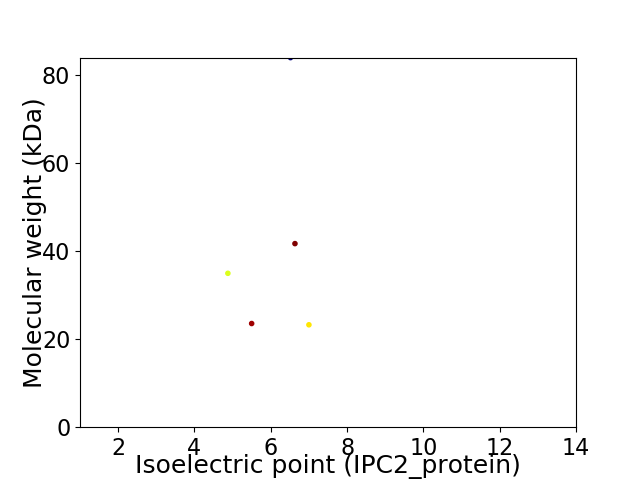
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C9X3W9|C9X3W9_9POLY Minor capsid protein VP2 OS=Sumatran orang-utan polyomavirus OX=1604875 GN=VP3 PE=3 SV=1
MM1 pKa = 7.49GGVISLIVDD10 pKa = 4.19IIEE13 pKa = 4.01LTAEE17 pKa = 3.97VTAATGFTVEE27 pKa = 5.01AIINGEE33 pKa = 3.69ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLSGVEE55 pKa = 4.03ALASLGITAEE65 pKa = 4.01QYY67 pKa = 10.29TFLTSVPGMLSNAVGLGVVFQTVSGASALVTAGIATSYY105 pKa = 10.37AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMALIPWRR122 pKa = 11.84DD123 pKa = 3.07PDD125 pKa = 3.8YY126 pKa = 11.55YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.67AIDD142 pKa = 4.39VITNWPGSLFHH153 pKa = 7.48SIGRR157 pKa = 11.84YY158 pKa = 8.43IWDD161 pKa = 3.58SVTSEE166 pKa = 3.54ARR168 pKa = 11.84GQIGHH173 pKa = 7.76ASNQVIVRR181 pKa = 11.84GTYY184 pKa = 10.37AFQDD188 pKa = 3.34SLARR192 pKa = 11.84ILEE195 pKa = 3.92NARR198 pKa = 11.84WVVQAGPTNVYY209 pKa = 10.7NFLDD213 pKa = 3.38SYY215 pKa = 11.2YY216 pKa = 10.71RR217 pKa = 11.84EE218 pKa = 4.43LPPINPAQARR228 pKa = 11.84QMYY231 pKa = 9.76RR232 pKa = 11.84RR233 pKa = 11.84IGQKK237 pKa = 10.04YY238 pKa = 8.62RR239 pKa = 11.84MPRR242 pKa = 11.84EE243 pKa = 3.57AEE245 pKa = 3.61IPDD248 pKa = 3.88RR249 pKa = 11.84YY250 pKa = 10.5NLEE253 pKa = 4.08SSEE256 pKa = 4.41SEE258 pKa = 3.74SGEE261 pKa = 3.86TVEE264 pKa = 5.8FYY266 pKa = 10.85KK267 pKa = 10.99HH268 pKa = 6.39PGGANQRR275 pKa = 11.84VTPDD279 pKa = 2.51WMLPLILGLYY289 pKa = 10.53GDD291 pKa = 4.96ITPTWDD297 pKa = 3.25KK298 pKa = 10.81YY299 pKa = 6.76VHH301 pKa = 5.8EE302 pKa = 4.51VEE304 pKa = 5.83EE305 pKa = 4.21EE306 pKa = 4.26DD307 pKa = 4.0EE308 pKa = 4.22LQKK311 pKa = 10.83KK312 pKa = 8.7KK313 pKa = 10.46RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84LL317 pKa = 3.37
MM1 pKa = 7.49GGVISLIVDD10 pKa = 4.19IIEE13 pKa = 4.01LTAEE17 pKa = 3.97VTAATGFTVEE27 pKa = 5.01AIINGEE33 pKa = 3.69ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLSGVEE55 pKa = 4.03ALASLGITAEE65 pKa = 4.01QYY67 pKa = 10.29TFLTSVPGMLSNAVGLGVVFQTVSGASALVTAGIATSYY105 pKa = 10.37AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMALIPWRR122 pKa = 11.84DD123 pKa = 3.07PDD125 pKa = 3.8YY126 pKa = 11.55YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.67AIDD142 pKa = 4.39VITNWPGSLFHH153 pKa = 7.48SIGRR157 pKa = 11.84YY158 pKa = 8.43IWDD161 pKa = 3.58SVTSEE166 pKa = 3.54ARR168 pKa = 11.84GQIGHH173 pKa = 7.76ASNQVIVRR181 pKa = 11.84GTYY184 pKa = 10.37AFQDD188 pKa = 3.34SLARR192 pKa = 11.84ILEE195 pKa = 3.92NARR198 pKa = 11.84WVVQAGPTNVYY209 pKa = 10.7NFLDD213 pKa = 3.38SYY215 pKa = 11.2YY216 pKa = 10.71RR217 pKa = 11.84EE218 pKa = 4.43LPPINPAQARR228 pKa = 11.84QMYY231 pKa = 9.76RR232 pKa = 11.84RR233 pKa = 11.84IGQKK237 pKa = 10.04YY238 pKa = 8.62RR239 pKa = 11.84MPRR242 pKa = 11.84EE243 pKa = 3.57AEE245 pKa = 3.61IPDD248 pKa = 3.88RR249 pKa = 11.84YY250 pKa = 10.5NLEE253 pKa = 4.08SSEE256 pKa = 4.41SEE258 pKa = 3.74SGEE261 pKa = 3.86TVEE264 pKa = 5.8FYY266 pKa = 10.85KK267 pKa = 10.99HH268 pKa = 6.39PGGANQRR275 pKa = 11.84VTPDD279 pKa = 2.51WMLPLILGLYY289 pKa = 10.53GDD291 pKa = 4.96ITPTWDD297 pKa = 3.25KK298 pKa = 10.81YY299 pKa = 6.76VHH301 pKa = 5.8EE302 pKa = 4.51VEE304 pKa = 5.83EE305 pKa = 4.21EE306 pKa = 4.26DD307 pKa = 4.0EE308 pKa = 4.22LQKK311 pKa = 10.83KK312 pKa = 8.7KK313 pKa = 10.46RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84LL317 pKa = 3.37
Molecular weight: 34.98 kDa
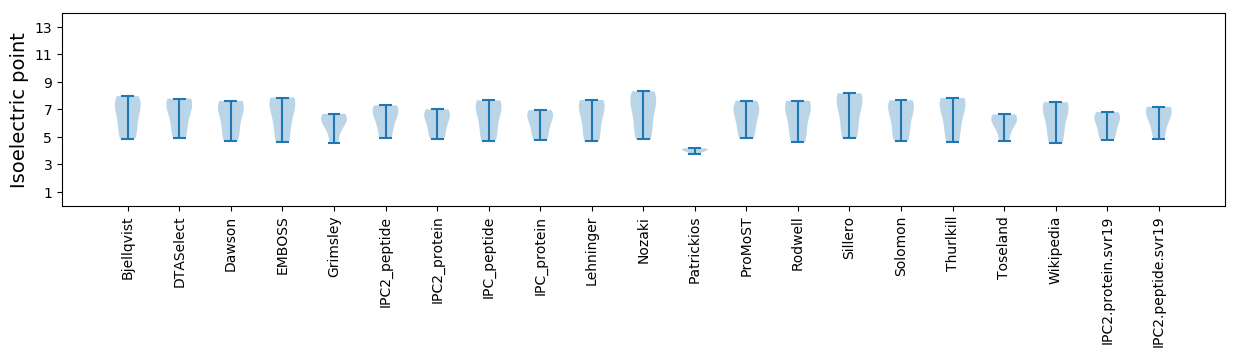
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9X3X2|C9X3X2_9POLY Small T antigen OS=Sumatran orang-utan polyomavirus OX=1604875 GN=small T PE=4 SV=1
MM1 pKa = 7.92DD2 pKa = 6.35KK3 pKa = 10.61ILTKK7 pKa = 7.75QQRR10 pKa = 11.84KK11 pKa = 7.55EE12 pKa = 4.17LIVLLDD18 pKa = 4.45IEE20 pKa = 4.49ACDD23 pKa = 3.64YY24 pKa = 11.64GNYY27 pKa = 10.49SIMKK31 pKa = 8.03QQYY34 pKa = 10.42KK35 pKa = 10.27KK36 pKa = 9.56MCLIYY41 pKa = 10.7HH42 pKa = 6.92PDD44 pKa = 3.33KK45 pKa = 11.58GGDD48 pKa = 3.62GEE50 pKa = 5.38KK51 pKa = 9.46MRR53 pKa = 11.84KK54 pKa = 9.29LNSLWQAFNTEE65 pKa = 4.02LLEE68 pKa = 4.44IRR70 pKa = 11.84DD71 pKa = 3.73NRR73 pKa = 11.84FDD75 pKa = 3.76MEE77 pKa = 4.35QVRR80 pKa = 11.84QMDD83 pKa = 3.35IDD85 pKa = 3.59VWDD88 pKa = 4.81DD89 pKa = 3.5LLTVKK94 pKa = 10.32EE95 pKa = 4.1ALDD98 pKa = 4.01DD99 pKa = 4.1FEE101 pKa = 6.77HH102 pKa = 7.31IFVKK106 pKa = 10.23VVPSCFNRR114 pKa = 11.84VAVCKK119 pKa = 9.86CKK121 pKa = 10.74CIVCKK126 pKa = 10.44LRR128 pKa = 11.84QQHH131 pKa = 7.0LIIKK135 pKa = 6.5EE136 pKa = 4.04HH137 pKa = 6.05KK138 pKa = 9.99RR139 pKa = 11.84CVLWGEE145 pKa = 4.4CFCHH149 pKa = 5.97KK150 pKa = 10.46CYY152 pKa = 11.0VEE154 pKa = 4.12WYY156 pKa = 8.85GLSGTEE162 pKa = 4.68QHH164 pKa = 6.79LIWWKK169 pKa = 10.73KK170 pKa = 9.92IIQDD174 pKa = 3.6TPFIFMRR181 pKa = 11.84LNTQDD186 pKa = 3.34TLKK189 pKa = 10.75PSLIYY194 pKa = 10.71
MM1 pKa = 7.92DD2 pKa = 6.35KK3 pKa = 10.61ILTKK7 pKa = 7.75QQRR10 pKa = 11.84KK11 pKa = 7.55EE12 pKa = 4.17LIVLLDD18 pKa = 4.45IEE20 pKa = 4.49ACDD23 pKa = 3.64YY24 pKa = 11.64GNYY27 pKa = 10.49SIMKK31 pKa = 8.03QQYY34 pKa = 10.42KK35 pKa = 10.27KK36 pKa = 9.56MCLIYY41 pKa = 10.7HH42 pKa = 6.92PDD44 pKa = 3.33KK45 pKa = 11.58GGDD48 pKa = 3.62GEE50 pKa = 5.38KK51 pKa = 9.46MRR53 pKa = 11.84KK54 pKa = 9.29LNSLWQAFNTEE65 pKa = 4.02LLEE68 pKa = 4.44IRR70 pKa = 11.84DD71 pKa = 3.73NRR73 pKa = 11.84FDD75 pKa = 3.76MEE77 pKa = 4.35QVRR80 pKa = 11.84QMDD83 pKa = 3.35IDD85 pKa = 3.59VWDD88 pKa = 4.81DD89 pKa = 3.5LLTVKK94 pKa = 10.32EE95 pKa = 4.1ALDD98 pKa = 4.01DD99 pKa = 4.1FEE101 pKa = 6.77HH102 pKa = 7.31IFVKK106 pKa = 10.23VVPSCFNRR114 pKa = 11.84VAVCKK119 pKa = 9.86CKK121 pKa = 10.74CIVCKK126 pKa = 10.44LRR128 pKa = 11.84QQHH131 pKa = 7.0LIIKK135 pKa = 6.5EE136 pKa = 4.04HH137 pKa = 6.05KK138 pKa = 9.99RR139 pKa = 11.84CVLWGEE145 pKa = 4.4CFCHH149 pKa = 5.97KK150 pKa = 10.46CYY152 pKa = 11.0VEE154 pKa = 4.12WYY156 pKa = 8.85GLSGTEE162 pKa = 4.68QHH164 pKa = 6.79LIWWKK169 pKa = 10.73KK170 pKa = 9.92IIQDD174 pKa = 3.6TPFIFMRR181 pKa = 11.84LNTQDD186 pKa = 3.34TLKK189 pKa = 10.75PSLIYY194 pKa = 10.71
Molecular weight: 23.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1828 |
194 |
735 |
365.6 |
41.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.853 ± 0.864 | 2.681 ± 0.831 |
5.635 ± 0.403 | 6.729 ± 0.345 |
4.431 ± 0.711 | 5.744 ± 0.732 |
1.641 ± 0.323 | 5.197 ± 0.814 |
6.783 ± 1.346 | 9.136 ± 0.668 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.681 ± 0.248 | 4.486 ± 0.333 |
5.47 ± 0.603 | 4.431 ± 0.289 |
4.869 ± 0.627 | 7.002 ± 0.715 |
5.525 ± 0.744 | 6.127 ± 0.876 |
1.477 ± 0.417 | 4.103 ± 0.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
