
Hesseltinella vesiculosa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Cunninghamellaceae; Hesseltinella
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
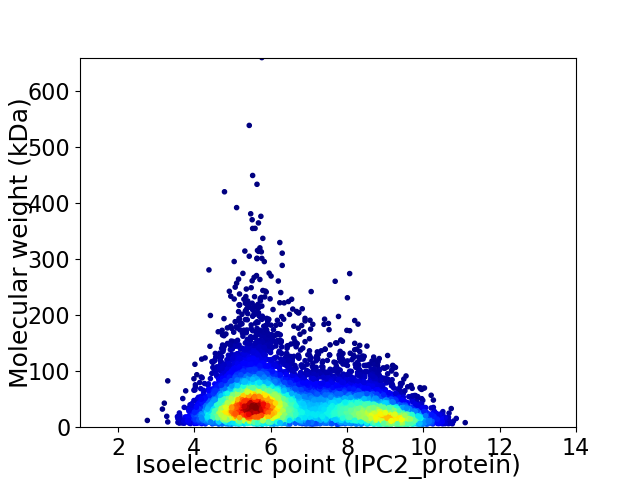
Virtual 2D-PAGE plot for 11058 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X2GUL2|A0A1X2GUL2_9FUNG Phospholipid-transporting ATPase OS=Hesseltinella vesiculosa OX=101127 GN=DM01DRAFT_1280350 PE=3 SV=1
MM1 pKa = 7.72RR2 pKa = 11.84GEE4 pKa = 5.03CMFFCYY10 pKa = 9.69QCQKK14 pKa = 10.89KK15 pKa = 10.19VGTIATSGADD25 pKa = 3.7TRR27 pKa = 11.84CSEE30 pKa = 4.28CLSVNIDD37 pKa = 3.71RR38 pKa = 11.84IGEE41 pKa = 4.12HH42 pKa = 6.61SDD44 pKa = 2.59WFYY47 pKa = 11.65DD48 pKa = 4.24EE49 pKa = 6.05SSQEE53 pKa = 4.0DD54 pKa = 4.03NDD56 pKa = 4.01RR57 pKa = 11.84DD58 pKa = 3.82DD59 pKa = 5.59YY60 pKa = 11.97NDD62 pKa = 4.0EE63 pKa = 4.13EE64 pKa = 4.97CEE66 pKa = 4.26SANEE70 pKa = 3.69QDD72 pKa = 3.7AAYY75 pKa = 10.72NDD77 pKa = 4.42LSHH80 pKa = 6.94LMSTVSVSSDD90 pKa = 2.85PGTDD94 pKa = 3.92LDD96 pKa = 3.98HH97 pKa = 7.15QEE99 pKa = 4.28NDD101 pKa = 4.48SPDD104 pKa = 3.62EE105 pKa = 4.98DD106 pKa = 4.77DD107 pKa = 5.95DD108 pKa = 5.43EE109 pKa = 4.9PVSDD113 pKa = 5.7DD114 pKa = 6.39NDD116 pKa = 4.21DD117 pKa = 3.09QWQDD121 pKa = 2.86MDD123 pKa = 4.81EE124 pKa = 4.73DD125 pKa = 4.93DD126 pKa = 5.57NLDD129 pKa = 3.48SDD131 pKa = 4.79EE132 pKa = 4.44EE133 pKa = 4.75DD134 pKa = 3.4EE135 pKa = 4.52AASPFHH141 pKa = 6.81LCPTHH146 pKa = 8.05ASMFAAAFSQAAQTATFHH164 pKa = 6.27TSDD167 pKa = 3.46PRR169 pKa = 11.84LRR171 pKa = 11.84LADD174 pKa = 4.06FMHH177 pKa = 7.17RR178 pKa = 11.84LYY180 pKa = 11.34SLDD183 pKa = 4.93DD184 pKa = 4.81DD185 pKa = 5.52DD186 pKa = 7.57DD187 pKa = 4.48YY188 pKa = 12.09FEE190 pKa = 6.51DD191 pKa = 5.19SDD193 pKa = 5.19YY194 pKa = 11.82DD195 pKa = 4.62DD196 pKa = 6.71DD197 pKa = 7.0DD198 pKa = 7.28DD199 pKa = 6.32EE200 pKa = 7.91DD201 pKa = 6.59DD202 pKa = 5.16DD203 pKa = 6.33DD204 pKa = 5.96EE205 pKa = 5.46EE206 pKa = 6.0EE207 pKa = 5.01EE208 pKa = 5.88DD209 pKa = 5.14DD210 pKa = 5.79DD211 pKa = 6.99DD212 pKa = 7.69DD213 pKa = 5.28IFADD217 pKa = 3.76EE218 pKa = 5.58DD219 pKa = 4.25GVSDD223 pKa = 5.47YY224 pKa = 11.97SNAWDD229 pKa = 3.55QNSNGDD235 pKa = 3.37SHH237 pKa = 8.04EE238 pKa = 5.28DD239 pKa = 3.35IEE241 pKa = 4.94EE242 pKa = 4.38LDD244 pKa = 4.62SLDD247 pKa = 4.39DD248 pKa = 5.29DD249 pKa = 4.33YY250 pKa = 12.11GEE252 pKa = 4.33MGNLGGFDD260 pKa = 3.82VDD262 pKa = 4.14TDD264 pKa = 5.35LLDD267 pKa = 4.21LLNNEE272 pKa = 5.02DD273 pKa = 4.33TDD275 pKa = 4.36DD276 pKa = 3.96MAWDD280 pKa = 3.84EE281 pKa = 4.52LAPSYY286 pKa = 10.72EE287 pKa = 4.23VLGTTLMADD296 pKa = 4.81RR297 pKa = 11.84SDD299 pKa = 3.77QDD301 pKa = 3.45DD302 pKa = 4.76GYY304 pKa = 10.62LGSVLEE310 pKa = 4.49VVHH313 pKa = 6.89RR314 pKa = 11.84EE315 pKa = 4.03YY316 pKa = 11.12EE317 pKa = 3.97PGPFEE322 pKa = 4.06EE323 pKa = 6.01HH324 pKa = 7.16PDD326 pKa = 3.66FLNGSIYY333 pKa = 10.64YY334 pKa = 9.69EE335 pKa = 4.1EE336 pKa = 4.13EE337 pKa = 4.12VVTRR341 pKa = 11.84RR342 pKa = 11.84HH343 pKa = 4.23QSRR346 pKa = 11.84FLRR349 pKa = 11.84LRR351 pKa = 11.84LRR353 pKa = 11.84HH354 pKa = 6.28IYY356 pKa = 9.06STMEE360 pKa = 4.06TNSEE364 pKa = 4.1AEE366 pKa = 4.37QPRR369 pKa = 11.84HH370 pKa = 5.84IPFEE374 pKa = 4.25NLSSRR379 pKa = 11.84TLHH382 pKa = 6.72DD383 pKa = 3.25QDD385 pKa = 4.44HH386 pKa = 6.61LVTASVDD393 pKa = 3.93CIICQEE399 pKa = 4.39TYY401 pKa = 8.08QTGKK405 pKa = 10.75LITKK409 pKa = 7.27MPCGHH414 pKa = 7.1EE415 pKa = 4.0YY416 pKa = 10.2HH417 pKa = 7.38DD418 pKa = 3.41EE419 pKa = 4.73CIRR422 pKa = 11.84HH423 pKa = 5.56WLNHH427 pKa = 6.54NITCPVCRR435 pKa = 11.84RR436 pKa = 11.84SLQEE440 pKa = 3.49VDD442 pKa = 3.86SVVSS446 pKa = 3.57
MM1 pKa = 7.72RR2 pKa = 11.84GEE4 pKa = 5.03CMFFCYY10 pKa = 9.69QCQKK14 pKa = 10.89KK15 pKa = 10.19VGTIATSGADD25 pKa = 3.7TRR27 pKa = 11.84CSEE30 pKa = 4.28CLSVNIDD37 pKa = 3.71RR38 pKa = 11.84IGEE41 pKa = 4.12HH42 pKa = 6.61SDD44 pKa = 2.59WFYY47 pKa = 11.65DD48 pKa = 4.24EE49 pKa = 6.05SSQEE53 pKa = 4.0DD54 pKa = 4.03NDD56 pKa = 4.01RR57 pKa = 11.84DD58 pKa = 3.82DD59 pKa = 5.59YY60 pKa = 11.97NDD62 pKa = 4.0EE63 pKa = 4.13EE64 pKa = 4.97CEE66 pKa = 4.26SANEE70 pKa = 3.69QDD72 pKa = 3.7AAYY75 pKa = 10.72NDD77 pKa = 4.42LSHH80 pKa = 6.94LMSTVSVSSDD90 pKa = 2.85PGTDD94 pKa = 3.92LDD96 pKa = 3.98HH97 pKa = 7.15QEE99 pKa = 4.28NDD101 pKa = 4.48SPDD104 pKa = 3.62EE105 pKa = 4.98DD106 pKa = 4.77DD107 pKa = 5.95DD108 pKa = 5.43EE109 pKa = 4.9PVSDD113 pKa = 5.7DD114 pKa = 6.39NDD116 pKa = 4.21DD117 pKa = 3.09QWQDD121 pKa = 2.86MDD123 pKa = 4.81EE124 pKa = 4.73DD125 pKa = 4.93DD126 pKa = 5.57NLDD129 pKa = 3.48SDD131 pKa = 4.79EE132 pKa = 4.44EE133 pKa = 4.75DD134 pKa = 3.4EE135 pKa = 4.52AASPFHH141 pKa = 6.81LCPTHH146 pKa = 8.05ASMFAAAFSQAAQTATFHH164 pKa = 6.27TSDD167 pKa = 3.46PRR169 pKa = 11.84LRR171 pKa = 11.84LADD174 pKa = 4.06FMHH177 pKa = 7.17RR178 pKa = 11.84LYY180 pKa = 11.34SLDD183 pKa = 4.93DD184 pKa = 4.81DD185 pKa = 5.52DD186 pKa = 7.57DD187 pKa = 4.48YY188 pKa = 12.09FEE190 pKa = 6.51DD191 pKa = 5.19SDD193 pKa = 5.19YY194 pKa = 11.82DD195 pKa = 4.62DD196 pKa = 6.71DD197 pKa = 7.0DD198 pKa = 7.28DD199 pKa = 6.32EE200 pKa = 7.91DD201 pKa = 6.59DD202 pKa = 5.16DD203 pKa = 6.33DD204 pKa = 5.96EE205 pKa = 5.46EE206 pKa = 6.0EE207 pKa = 5.01EE208 pKa = 5.88DD209 pKa = 5.14DD210 pKa = 5.79DD211 pKa = 6.99DD212 pKa = 7.69DD213 pKa = 5.28IFADD217 pKa = 3.76EE218 pKa = 5.58DD219 pKa = 4.25GVSDD223 pKa = 5.47YY224 pKa = 11.97SNAWDD229 pKa = 3.55QNSNGDD235 pKa = 3.37SHH237 pKa = 8.04EE238 pKa = 5.28DD239 pKa = 3.35IEE241 pKa = 4.94EE242 pKa = 4.38LDD244 pKa = 4.62SLDD247 pKa = 4.39DD248 pKa = 5.29DD249 pKa = 4.33YY250 pKa = 12.11GEE252 pKa = 4.33MGNLGGFDD260 pKa = 3.82VDD262 pKa = 4.14TDD264 pKa = 5.35LLDD267 pKa = 4.21LLNNEE272 pKa = 5.02DD273 pKa = 4.33TDD275 pKa = 4.36DD276 pKa = 3.96MAWDD280 pKa = 3.84EE281 pKa = 4.52LAPSYY286 pKa = 10.72EE287 pKa = 4.23VLGTTLMADD296 pKa = 4.81RR297 pKa = 11.84SDD299 pKa = 3.77QDD301 pKa = 3.45DD302 pKa = 4.76GYY304 pKa = 10.62LGSVLEE310 pKa = 4.49VVHH313 pKa = 6.89RR314 pKa = 11.84EE315 pKa = 4.03YY316 pKa = 11.12EE317 pKa = 3.97PGPFEE322 pKa = 4.06EE323 pKa = 6.01HH324 pKa = 7.16PDD326 pKa = 3.66FLNGSIYY333 pKa = 10.64YY334 pKa = 9.69EE335 pKa = 4.1EE336 pKa = 4.13EE337 pKa = 4.12VVTRR341 pKa = 11.84RR342 pKa = 11.84HH343 pKa = 4.23QSRR346 pKa = 11.84FLRR349 pKa = 11.84LRR351 pKa = 11.84LRR353 pKa = 11.84HH354 pKa = 6.28IYY356 pKa = 9.06STMEE360 pKa = 4.06TNSEE364 pKa = 4.1AEE366 pKa = 4.37QPRR369 pKa = 11.84HH370 pKa = 5.84IPFEE374 pKa = 4.25NLSSRR379 pKa = 11.84TLHH382 pKa = 6.72DD383 pKa = 3.25QDD385 pKa = 4.44HH386 pKa = 6.61LVTASVDD393 pKa = 3.93CIICQEE399 pKa = 4.39TYY401 pKa = 8.08QTGKK405 pKa = 10.75LITKK409 pKa = 7.27MPCGHH414 pKa = 7.1EE415 pKa = 4.0YY416 pKa = 10.2HH417 pKa = 7.38DD418 pKa = 3.41EE419 pKa = 4.73CIRR422 pKa = 11.84HH423 pKa = 5.56WLNHH427 pKa = 6.54NITCPVCRR435 pKa = 11.84RR436 pKa = 11.84SLQEE440 pKa = 3.49VDD442 pKa = 3.86SVVSS446 pKa = 3.57
Molecular weight: 51.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X2GIR6|A0A1X2GIR6_9FUNG Elongation of fatty acids protein OS=Hesseltinella vesiculosa OX=101127 GN=DM01DRAFT_1407422 PE=3 SV=1
NN1 pKa = 6.58QKK3 pKa = 10.46SAVSQRR9 pKa = 11.84KK10 pKa = 8.66SAVSQRR16 pKa = 11.84KK17 pKa = 8.66SAVSQRR23 pKa = 11.84KK24 pKa = 8.5SAGANQKK31 pKa = 10.35SAVSQRR37 pKa = 11.84KK38 pKa = 8.66SAVSQRR44 pKa = 11.84KK45 pKa = 8.5SAGANQKK52 pKa = 7.84TSKK55 pKa = 10.34KK56 pKa = 10.26SAGANQKK63 pKa = 10.35SAVSQRR69 pKa = 11.84KK70 pKa = 8.66SAVSQRR76 pKa = 11.84KK77 pKa = 8.68SAVSQQKK84 pKa = 9.82NAVSQRR90 pKa = 11.84KK91 pKa = 8.48SAGANQKK98 pKa = 10.35SAVSQRR104 pKa = 11.84KK105 pKa = 8.68SAVSQQKK112 pKa = 9.75KK113 pKa = 9.88CRR115 pKa = 11.84SQSKK119 pKa = 9.41KK120 pKa = 10.11CRR122 pKa = 11.84SQRR125 pKa = 11.84KK126 pKa = 8.48SAVSQRR132 pKa = 11.84KK133 pKa = 8.68SAVSQQKK140 pKa = 9.95IPGKK144 pKa = 9.72FQQ146 pKa = 3.29
NN1 pKa = 6.58QKK3 pKa = 10.46SAVSQRR9 pKa = 11.84KK10 pKa = 8.66SAVSQRR16 pKa = 11.84KK17 pKa = 8.66SAVSQRR23 pKa = 11.84KK24 pKa = 8.5SAGANQKK31 pKa = 10.35SAVSQRR37 pKa = 11.84KK38 pKa = 8.66SAVSQRR44 pKa = 11.84KK45 pKa = 8.5SAGANQKK52 pKa = 7.84TSKK55 pKa = 10.34KK56 pKa = 10.26SAGANQKK63 pKa = 10.35SAVSQRR69 pKa = 11.84KK70 pKa = 8.66SAVSQRR76 pKa = 11.84KK77 pKa = 8.68SAVSQQKK84 pKa = 9.82NAVSQRR90 pKa = 11.84KK91 pKa = 8.48SAGANQKK98 pKa = 10.35SAVSQRR104 pKa = 11.84KK105 pKa = 8.68SAVSQQKK112 pKa = 9.75KK113 pKa = 9.88CRR115 pKa = 11.84SQSKK119 pKa = 9.41KK120 pKa = 10.11CRR122 pKa = 11.84SQRR125 pKa = 11.84KK126 pKa = 8.48SAVSQRR132 pKa = 11.84KK133 pKa = 8.68SAVSQQKK140 pKa = 9.95IPGKK144 pKa = 9.72FQQ146 pKa = 3.29
Molecular weight: 15.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4440516 |
49 |
6093 |
401.6 |
45.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.583 ± 0.022 | 1.475 ± 0.011 |
6.154 ± 0.022 | 5.115 ± 0.026 |
3.896 ± 0.017 | 5.158 ± 0.023 |
3.059 ± 0.016 | 5.002 ± 0.019 |
5.167 ± 0.025 | 9.848 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.01 | 3.915 ± 0.015 |
5.897 ± 0.03 | 5.096 ± 0.024 |
5.247 ± 0.019 | 8.069 ± 0.034 |
6.204 ± 0.021 | 6.12 ± 0.022 |
1.28 ± 0.009 | 3.082 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
