
Dermatophagoides pteronyssinus (European house dust mite)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Chelicerata; Arachnida; Acari; Acariformes; Sarcoptiformes; Astigmata; Psoroptidia; Analgoidea; Pyroglyphidae; Dermatophagoidinae; Dermatophagoides
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
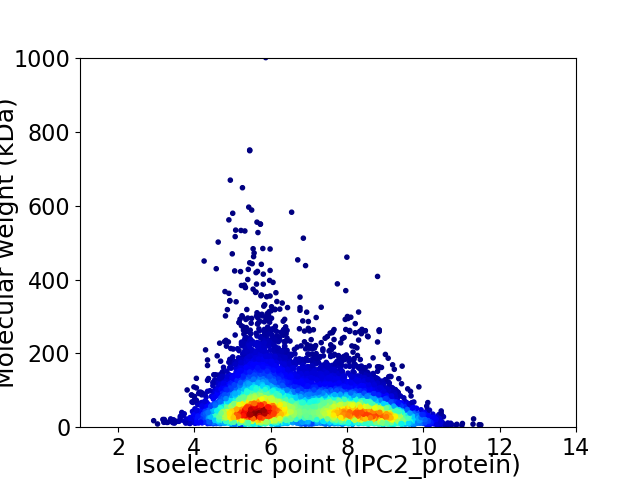
Virtual 2D-PAGE plot for 12480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P6Y1C1|A0A6P6Y1C1_DERPT eIF-2B GDP-GTP exchange factor subunit alpha OS=Dermatophagoides pteronyssinus OX=6956 GN=LOC113792416 PE=3 SV=1
MM1 pKa = 7.76SDD3 pKa = 3.99SDD5 pKa = 5.01DD6 pKa = 4.79FSDD9 pKa = 5.28DD10 pKa = 3.43DD11 pKa = 5.2HH12 pKa = 9.57SFDD15 pKa = 4.38SHH17 pKa = 7.21NDD19 pKa = 3.15SLDD22 pKa = 3.94GYY24 pKa = 11.13DD25 pKa = 5.56SDD27 pKa = 5.97DD28 pKa = 4.33FSNDD32 pKa = 2.41DD33 pKa = 3.64HH34 pKa = 7.07TFDD37 pKa = 4.22SHH39 pKa = 7.07NNSLDD44 pKa = 3.21GYY46 pKa = 10.04YY47 pKa = 10.76DD48 pKa = 3.9SDD50 pKa = 3.9VLNCSDD56 pKa = 3.92VDD58 pKa = 3.87DD59 pKa = 5.85ANFSDD64 pKa = 5.14DD65 pKa = 4.0HH66 pKa = 8.79ASDD69 pKa = 4.96DD70 pKa = 4.11LLFDD74 pKa = 3.94EE75 pKa = 5.02EE76 pKa = 4.97QNYY79 pKa = 10.44SDD81 pKa = 3.36NDD83 pKa = 3.42NRR85 pKa = 11.84IEE87 pKa = 4.33EE88 pKa = 4.22LDD90 pKa = 3.75EE91 pKa = 5.0NNEE94 pKa = 4.02QQHH97 pKa = 6.48LEE99 pKa = 3.98NEE101 pKa = 4.59MINDD105 pKa = 3.73DD106 pKa = 3.85TSSEE110 pKa = 4.12EE111 pKa = 3.65IEE113 pKa = 4.37EE114 pKa = 4.22NSNHH118 pKa = 5.95SVISDD123 pKa = 3.61DD124 pKa = 4.67DD125 pKa = 4.06NEE127 pKa = 4.41EE128 pKa = 3.97NVLDD132 pKa = 4.51DD133 pKa = 5.31NISEE137 pKa = 4.4YY138 pKa = 11.34VPDD141 pKa = 4.0TDD143 pKa = 6.4DD144 pKa = 5.54YY145 pKa = 12.1DD146 pKa = 6.73DD147 pKa = 6.7DD148 pKa = 7.47DD149 pKa = 7.49DD150 pKa = 7.6DD151 pKa = 7.71DD152 pKa = 7.3DD153 pKa = 6.78DD154 pKa = 6.14DD155 pKa = 6.2SSNQLYY161 pKa = 10.24TNFLMSLDD169 pKa = 3.88HH170 pKa = 5.56QTDD173 pKa = 3.7YY174 pKa = 11.36NGPVAVDD181 pKa = 3.28QQSNDD186 pKa = 3.15KK187 pKa = 10.51KK188 pKa = 11.07YY189 pKa = 11.07KK190 pKa = 9.06NQEE193 pKa = 3.89TQTDD197 pKa = 3.84VTNLIVQNRR206 pKa = 11.84CSICLEE212 pKa = 4.21SFCPNKK218 pKa = 10.3DD219 pKa = 3.26HH220 pKa = 7.39YY221 pKa = 10.64VVCLPCGHH229 pKa = 7.56LFGKK233 pKa = 8.09TCIEE237 pKa = 3.79KK238 pKa = 9.63WLASSKK244 pKa = 10.58YY245 pKa = 9.8CPTCRR250 pKa = 11.84FHH252 pKa = 8.96AEE254 pKa = 3.34IGDD257 pKa = 3.89IIKK260 pKa = 10.39IFLNPIDD267 pKa = 4.22INLMKK272 pKa = 10.46QNCEE276 pKa = 3.82TKK278 pKa = 9.64FWRR281 pKa = 11.84DD282 pKa = 2.79KK283 pKa = 9.01TTEE286 pKa = 3.87LTLEE290 pKa = 3.99NLQLKK295 pKa = 9.15MEE297 pKa = 4.56LHH299 pKa = 5.87LAKK302 pKa = 10.67NKK304 pKa = 10.08IINQIDD310 pKa = 3.85FNEE313 pKa = 4.36KK314 pKa = 10.12
MM1 pKa = 7.76SDD3 pKa = 3.99SDD5 pKa = 5.01DD6 pKa = 4.79FSDD9 pKa = 5.28DD10 pKa = 3.43DD11 pKa = 5.2HH12 pKa = 9.57SFDD15 pKa = 4.38SHH17 pKa = 7.21NDD19 pKa = 3.15SLDD22 pKa = 3.94GYY24 pKa = 11.13DD25 pKa = 5.56SDD27 pKa = 5.97DD28 pKa = 4.33FSNDD32 pKa = 2.41DD33 pKa = 3.64HH34 pKa = 7.07TFDD37 pKa = 4.22SHH39 pKa = 7.07NNSLDD44 pKa = 3.21GYY46 pKa = 10.04YY47 pKa = 10.76DD48 pKa = 3.9SDD50 pKa = 3.9VLNCSDD56 pKa = 3.92VDD58 pKa = 3.87DD59 pKa = 5.85ANFSDD64 pKa = 5.14DD65 pKa = 4.0HH66 pKa = 8.79ASDD69 pKa = 4.96DD70 pKa = 4.11LLFDD74 pKa = 3.94EE75 pKa = 5.02EE76 pKa = 4.97QNYY79 pKa = 10.44SDD81 pKa = 3.36NDD83 pKa = 3.42NRR85 pKa = 11.84IEE87 pKa = 4.33EE88 pKa = 4.22LDD90 pKa = 3.75EE91 pKa = 5.0NNEE94 pKa = 4.02QQHH97 pKa = 6.48LEE99 pKa = 3.98NEE101 pKa = 4.59MINDD105 pKa = 3.73DD106 pKa = 3.85TSSEE110 pKa = 4.12EE111 pKa = 3.65IEE113 pKa = 4.37EE114 pKa = 4.22NSNHH118 pKa = 5.95SVISDD123 pKa = 3.61DD124 pKa = 4.67DD125 pKa = 4.06NEE127 pKa = 4.41EE128 pKa = 3.97NVLDD132 pKa = 4.51DD133 pKa = 5.31NISEE137 pKa = 4.4YY138 pKa = 11.34VPDD141 pKa = 4.0TDD143 pKa = 6.4DD144 pKa = 5.54YY145 pKa = 12.1DD146 pKa = 6.73DD147 pKa = 6.7DD148 pKa = 7.47DD149 pKa = 7.49DD150 pKa = 7.6DD151 pKa = 7.71DD152 pKa = 7.3DD153 pKa = 6.78DD154 pKa = 6.14DD155 pKa = 6.2SSNQLYY161 pKa = 10.24TNFLMSLDD169 pKa = 3.88HH170 pKa = 5.56QTDD173 pKa = 3.7YY174 pKa = 11.36NGPVAVDD181 pKa = 3.28QQSNDD186 pKa = 3.15KK187 pKa = 10.51KK188 pKa = 11.07YY189 pKa = 11.07KK190 pKa = 9.06NQEE193 pKa = 3.89TQTDD197 pKa = 3.84VTNLIVQNRR206 pKa = 11.84CSICLEE212 pKa = 4.21SFCPNKK218 pKa = 10.3DD219 pKa = 3.26HH220 pKa = 7.39YY221 pKa = 10.64VVCLPCGHH229 pKa = 7.56LFGKK233 pKa = 8.09TCIEE237 pKa = 3.79KK238 pKa = 9.63WLASSKK244 pKa = 10.58YY245 pKa = 9.8CPTCRR250 pKa = 11.84FHH252 pKa = 8.96AEE254 pKa = 3.34IGDD257 pKa = 3.89IIKK260 pKa = 10.39IFLNPIDD267 pKa = 4.22INLMKK272 pKa = 10.46QNCEE276 pKa = 3.82TKK278 pKa = 9.64FWRR281 pKa = 11.84DD282 pKa = 2.79KK283 pKa = 9.01TTEE286 pKa = 3.87LTLEE290 pKa = 3.99NLQLKK295 pKa = 9.15MEE297 pKa = 4.56LHH299 pKa = 5.87LAKK302 pKa = 10.67NKK304 pKa = 10.08IINQIDD310 pKa = 3.85FNEE313 pKa = 4.36KK314 pKa = 10.12
Molecular weight: 36.32 kDa
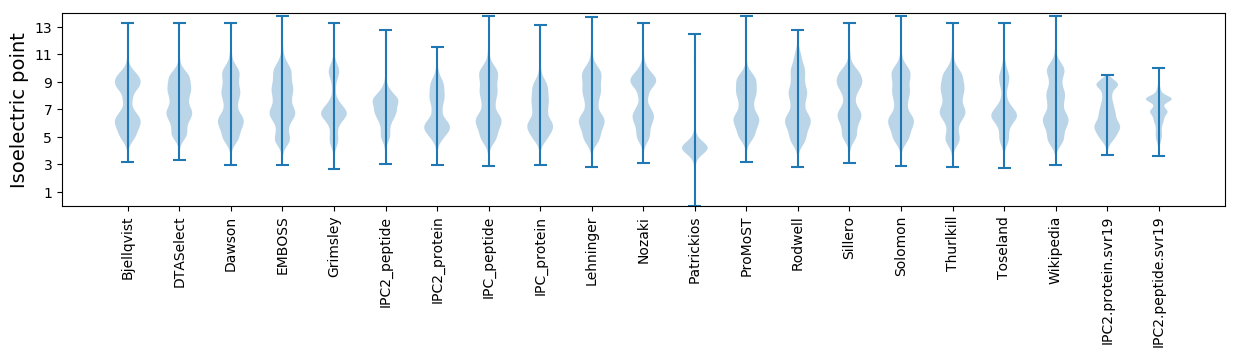
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P6Y7E7|A0A6P6Y7E7_DERPT DNA-directed RNA polymerase subunit OS=Dermatophagoides pteronyssinus OX=6956 GN=LOC113795351 PE=3 SV=1
RR1 pKa = 7.49LSRR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84RR9 pKa = 11.84GARR12 pKa = 11.84RR13 pKa = 11.84AAASRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84AAGRR26 pKa = 11.84RR27 pKa = 11.84SRR29 pKa = 11.84TRR31 pKa = 11.84GSRR34 pKa = 11.84APPRR38 pKa = 11.84STRR41 pKa = 11.84RR42 pKa = 11.84AASTATTNRR51 pKa = 11.84ASRR54 pKa = 11.84SRR56 pKa = 11.84ARR58 pKa = 11.84ARR60 pKa = 11.84FCSSSSRR67 pKa = 11.84SRR69 pKa = 11.84FSSSSS74 pKa = 3.05
RR1 pKa = 7.49LSRR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84RR9 pKa = 11.84GARR12 pKa = 11.84RR13 pKa = 11.84AAASRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84AAGRR26 pKa = 11.84RR27 pKa = 11.84SRR29 pKa = 11.84TRR31 pKa = 11.84GSRR34 pKa = 11.84APPRR38 pKa = 11.84STRR41 pKa = 11.84RR42 pKa = 11.84AASTATTNRR51 pKa = 11.84ASRR54 pKa = 11.84SRR56 pKa = 11.84ARR58 pKa = 11.84ARR60 pKa = 11.84FCSSSSRR67 pKa = 11.84SRR69 pKa = 11.84FSSSSS74 pKa = 3.05
Molecular weight: 8.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7101821 |
20 |
8940 |
569.1 |
64.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.491 ± 0.022 | 1.782 ± 0.017 |
5.838 ± 0.019 | 5.419 ± 0.026 |
4.312 ± 0.019 | 4.442 ± 0.023 |
2.875 ± 0.013 | 7.629 ± 0.023 |
6.176 ± 0.024 | 8.717 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.008 | 7.584 ± 0.028 |
4.224 ± 0.022 | 5.449 ± 0.026 |
4.675 ± 0.014 | 9.331 ± 0.035 |
5.895 ± 0.025 | 4.402 ± 0.016 |
0.927 ± 0.007 | 3.386 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
