
Avon-Heathcote Estuary associated circular virus 11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
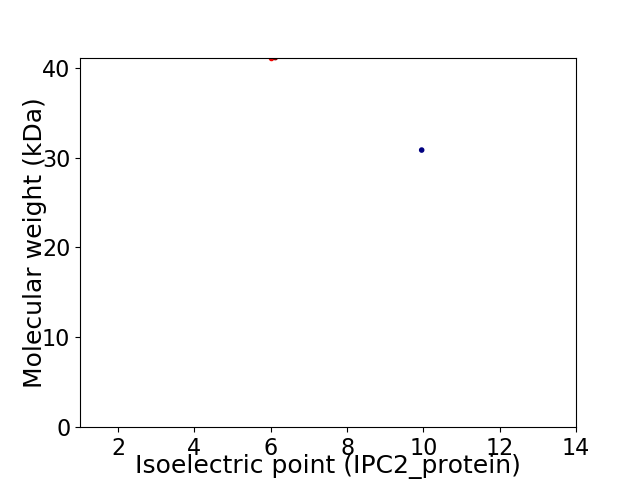
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
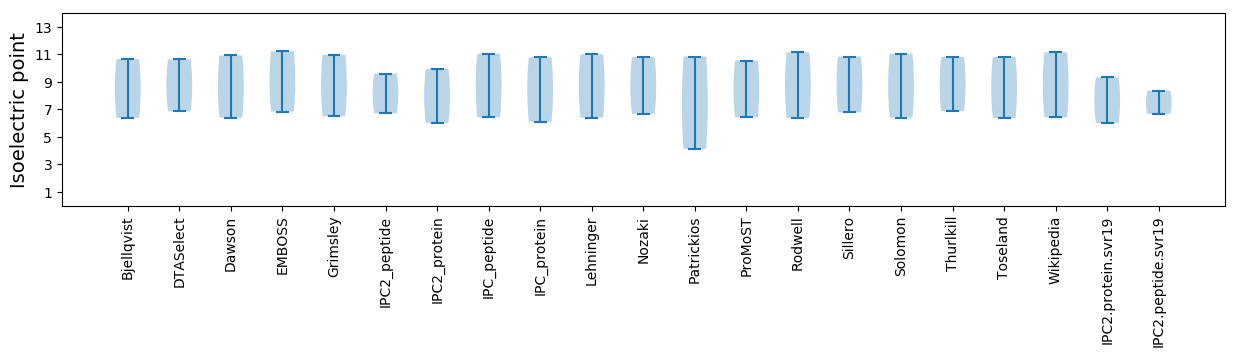
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IB17|A0A0C5IB17_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 11 OX=1618234 PE=3 SV=1
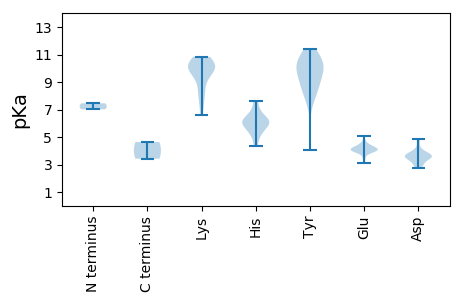
MM1 pKa = 7.46IAHH4 pKa = 6.14NRR6 pKa = 11.84AFFTISITGLKK17 pKa = 9.73GMCDD21 pKa = 2.88IIFFLFLSLAQMASPKK37 pKa = 10.0KK38 pKa = 7.67YY39 pKa = 10.04NRR41 pKa = 11.84MIFTINNPGPNRR53 pKa = 11.84PQFLEE58 pKa = 3.4QHH60 pKa = 5.2MAYY63 pKa = 9.29MIYY66 pKa = 10.37VLEE69 pKa = 5.07EE70 pKa = 4.13GDD72 pKa = 3.71QTHH75 pKa = 6.29TPHH78 pKa = 5.94LQGYY82 pKa = 8.62IRR84 pKa = 11.84WKK86 pKa = 9.74CRR88 pKa = 11.84KK89 pKa = 6.57TFKK92 pKa = 10.38SAEE95 pKa = 3.97RR96 pKa = 11.84LLGGRR101 pKa = 11.84AYY103 pKa = 11.05LEE105 pKa = 3.92IARR108 pKa = 11.84GTEE111 pKa = 4.05EE112 pKa = 4.71EE113 pKa = 3.99NQAYY117 pKa = 9.22CKK119 pKa = 10.41KK120 pKa = 10.32EE121 pKa = 3.96EE122 pKa = 4.35GVLEE126 pKa = 4.25PWQEE130 pKa = 3.59FGTYY134 pKa = 10.15DD135 pKa = 3.44GGSGIKK141 pKa = 10.05GRR143 pKa = 11.84RR144 pKa = 11.84TDD146 pKa = 3.66LEE148 pKa = 4.03AVAKK152 pKa = 9.78RR153 pKa = 11.84VRR155 pKa = 11.84EE156 pKa = 4.11GASARR161 pKa = 11.84TIAVEE166 pKa = 3.94HH167 pKa = 5.86TEE169 pKa = 4.23EE170 pKa = 4.53FIKK173 pKa = 10.79YY174 pKa = 8.83HH175 pKa = 6.22AGIEE179 pKa = 4.2RR180 pKa = 11.84AVYY183 pKa = 8.31MARR186 pKa = 11.84PEE188 pKa = 4.25PPAQRR193 pKa = 11.84QIEE196 pKa = 4.13MMIIWGPSGIGKK208 pKa = 7.22THH210 pKa = 6.11AVMMNDD216 pKa = 3.57GLSEE220 pKa = 4.18SGGVYY225 pKa = 10.08CVPTGNHH232 pKa = 6.41PWDD235 pKa = 3.66QYY237 pKa = 11.05EE238 pKa = 4.43GEE240 pKa = 4.26ATIFMDD246 pKa = 4.08EE247 pKa = 4.29FRR249 pKa = 11.84WEE251 pKa = 3.86HH252 pKa = 5.74WEE254 pKa = 4.0CTLMNRR260 pKa = 11.84ILDD263 pKa = 3.58KK264 pKa = 10.73WRR266 pKa = 11.84LQLPCRR272 pKa = 11.84YY273 pKa = 7.86MNKK276 pKa = 8.0MAAWTRR282 pKa = 11.84IIICTNQDD290 pKa = 2.76PMTWYY295 pKa = 9.82PNEE298 pKa = 4.03EE299 pKa = 4.11LPIRR303 pKa = 11.84DD304 pKa = 3.53ALRR307 pKa = 11.84RR308 pKa = 11.84RR309 pKa = 11.84LGTNCRR315 pKa = 11.84HH316 pKa = 4.31MTEE319 pKa = 4.49RR320 pKa = 11.84GQDD323 pKa = 3.53LAASPPSPDD332 pKa = 3.13FSSYY336 pKa = 10.3IPPAMRR342 pKa = 11.84VQDD345 pKa = 4.12SLDD348 pKa = 3.88DD349 pKa = 3.82ATVPASDD356 pKa = 3.45SS357 pKa = 3.42
MM1 pKa = 7.46IAHH4 pKa = 6.14NRR6 pKa = 11.84AFFTISITGLKK17 pKa = 9.73GMCDD21 pKa = 2.88IIFFLFLSLAQMASPKK37 pKa = 10.0KK38 pKa = 7.67YY39 pKa = 10.04NRR41 pKa = 11.84MIFTINNPGPNRR53 pKa = 11.84PQFLEE58 pKa = 3.4QHH60 pKa = 5.2MAYY63 pKa = 9.29MIYY66 pKa = 10.37VLEE69 pKa = 5.07EE70 pKa = 4.13GDD72 pKa = 3.71QTHH75 pKa = 6.29TPHH78 pKa = 5.94LQGYY82 pKa = 8.62IRR84 pKa = 11.84WKK86 pKa = 9.74CRR88 pKa = 11.84KK89 pKa = 6.57TFKK92 pKa = 10.38SAEE95 pKa = 3.97RR96 pKa = 11.84LLGGRR101 pKa = 11.84AYY103 pKa = 11.05LEE105 pKa = 3.92IARR108 pKa = 11.84GTEE111 pKa = 4.05EE112 pKa = 4.71EE113 pKa = 3.99NQAYY117 pKa = 9.22CKK119 pKa = 10.41KK120 pKa = 10.32EE121 pKa = 3.96EE122 pKa = 4.35GVLEE126 pKa = 4.25PWQEE130 pKa = 3.59FGTYY134 pKa = 10.15DD135 pKa = 3.44GGSGIKK141 pKa = 10.05GRR143 pKa = 11.84RR144 pKa = 11.84TDD146 pKa = 3.66LEE148 pKa = 4.03AVAKK152 pKa = 9.78RR153 pKa = 11.84VRR155 pKa = 11.84EE156 pKa = 4.11GASARR161 pKa = 11.84TIAVEE166 pKa = 3.94HH167 pKa = 5.86TEE169 pKa = 4.23EE170 pKa = 4.53FIKK173 pKa = 10.79YY174 pKa = 8.83HH175 pKa = 6.22AGIEE179 pKa = 4.2RR180 pKa = 11.84AVYY183 pKa = 8.31MARR186 pKa = 11.84PEE188 pKa = 4.25PPAQRR193 pKa = 11.84QIEE196 pKa = 4.13MMIIWGPSGIGKK208 pKa = 7.22THH210 pKa = 6.11AVMMNDD216 pKa = 3.57GLSEE220 pKa = 4.18SGGVYY225 pKa = 10.08CVPTGNHH232 pKa = 6.41PWDD235 pKa = 3.66QYY237 pKa = 11.05EE238 pKa = 4.43GEE240 pKa = 4.26ATIFMDD246 pKa = 4.08EE247 pKa = 4.29FRR249 pKa = 11.84WEE251 pKa = 3.86HH252 pKa = 5.74WEE254 pKa = 4.0CTLMNRR260 pKa = 11.84ILDD263 pKa = 3.58KK264 pKa = 10.73WRR266 pKa = 11.84LQLPCRR272 pKa = 11.84YY273 pKa = 7.86MNKK276 pKa = 8.0MAAWTRR282 pKa = 11.84IIICTNQDD290 pKa = 2.76PMTWYY295 pKa = 9.82PNEE298 pKa = 4.03EE299 pKa = 4.11LPIRR303 pKa = 11.84DD304 pKa = 3.53ALRR307 pKa = 11.84RR308 pKa = 11.84RR309 pKa = 11.84LGTNCRR315 pKa = 11.84HH316 pKa = 4.31MTEE319 pKa = 4.49RR320 pKa = 11.84GQDD323 pKa = 3.53LAASPPSPDD332 pKa = 3.13FSSYY336 pKa = 10.3IPPAMRR342 pKa = 11.84VQDD345 pKa = 4.12SLDD348 pKa = 3.88DD349 pKa = 3.82ATVPASDD356 pKa = 3.45SS357 pKa = 3.42
Molecular weight: 41.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IB17|A0A0C5IB17_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 11 OX=1618234 PE=3 SV=1
MM1 pKa = 7.01VYY3 pKa = 10.36KK4 pKa = 10.47RR5 pKa = 11.84RR6 pKa = 11.84SPFKK10 pKa = 10.19KK11 pKa = 10.0LSSRR15 pKa = 11.84PRR17 pKa = 11.84KK18 pKa = 8.36RR19 pKa = 11.84ARR21 pKa = 11.84YY22 pKa = 4.08TTKK25 pKa = 8.36TARR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84RR32 pKa = 11.84TIRR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 9.32RR38 pKa = 11.84RR39 pKa = 11.84GYY41 pKa = 8.81KK42 pKa = 9.83ARR44 pKa = 11.84AARR47 pKa = 11.84GRR49 pKa = 11.84TNNTVANMFGLRR61 pKa = 11.84ATTKK65 pKa = 10.01SVRR68 pKa = 11.84LVHH71 pKa = 6.97SITEE75 pKa = 4.03AFAPSLAAASSVAVTAFTRR94 pKa = 11.84TINNPLDD101 pKa = 3.67PMYY104 pKa = 10.3TAGSQYY110 pKa = 10.28ATGFKK115 pKa = 10.06EE116 pKa = 3.87LAANYY121 pKa = 8.55MYY123 pKa = 11.41YY124 pKa = 10.16EE125 pKa = 4.16VQKK128 pKa = 10.84ACLDD132 pKa = 2.97IWVRR136 pKa = 11.84QSANYY141 pKa = 7.59VTAASYY147 pKa = 10.66PDD149 pKa = 4.63LIIMAKK155 pKa = 10.36LNNDD159 pKa = 3.56WTWGDD164 pKa = 3.51IQDD167 pKa = 4.84LRR169 pKa = 11.84VDD171 pKa = 3.29QWAQLANIKK180 pKa = 8.88VARR183 pKa = 11.84LRR185 pKa = 11.84FHH187 pKa = 7.6PEE189 pKa = 3.08MTGAKK194 pKa = 9.53CHH196 pKa = 6.08IRR198 pKa = 11.84LWWHH202 pKa = 5.26SKK204 pKa = 10.77KK205 pKa = 9.06NTKK208 pKa = 9.39TDD210 pKa = 3.71DD211 pKa = 3.42NVAATTASPSTIQPVAVGAFSADD234 pKa = 3.21NSTVASLPAMTISSRR249 pKa = 11.84IRR251 pKa = 11.84FWTKK255 pKa = 9.93FSMLKK260 pKa = 10.47PLDD263 pKa = 3.78TDD265 pKa = 3.79TLQDD269 pKa = 2.93IMTT272 pKa = 4.63
MM1 pKa = 7.01VYY3 pKa = 10.36KK4 pKa = 10.47RR5 pKa = 11.84RR6 pKa = 11.84SPFKK10 pKa = 10.19KK11 pKa = 10.0LSSRR15 pKa = 11.84PRR17 pKa = 11.84KK18 pKa = 8.36RR19 pKa = 11.84ARR21 pKa = 11.84YY22 pKa = 4.08TTKK25 pKa = 8.36TARR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84RR32 pKa = 11.84TIRR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 9.32RR38 pKa = 11.84RR39 pKa = 11.84GYY41 pKa = 8.81KK42 pKa = 9.83ARR44 pKa = 11.84AARR47 pKa = 11.84GRR49 pKa = 11.84TNNTVANMFGLRR61 pKa = 11.84ATTKK65 pKa = 10.01SVRR68 pKa = 11.84LVHH71 pKa = 6.97SITEE75 pKa = 4.03AFAPSLAAASSVAVTAFTRR94 pKa = 11.84TINNPLDD101 pKa = 3.67PMYY104 pKa = 10.3TAGSQYY110 pKa = 10.28ATGFKK115 pKa = 10.06EE116 pKa = 3.87LAANYY121 pKa = 8.55MYY123 pKa = 11.41YY124 pKa = 10.16EE125 pKa = 4.16VQKK128 pKa = 10.84ACLDD132 pKa = 2.97IWVRR136 pKa = 11.84QSANYY141 pKa = 7.59VTAASYY147 pKa = 10.66PDD149 pKa = 4.63LIIMAKK155 pKa = 10.36LNNDD159 pKa = 3.56WTWGDD164 pKa = 3.51IQDD167 pKa = 4.84LRR169 pKa = 11.84VDD171 pKa = 3.29QWAQLANIKK180 pKa = 8.88VARR183 pKa = 11.84LRR185 pKa = 11.84FHH187 pKa = 7.6PEE189 pKa = 3.08MTGAKK194 pKa = 9.53CHH196 pKa = 6.08IRR198 pKa = 11.84LWWHH202 pKa = 5.26SKK204 pKa = 10.77KK205 pKa = 9.06NTKK208 pKa = 9.39TDD210 pKa = 3.71DD211 pKa = 3.42NVAATTASPSTIQPVAVGAFSADD234 pKa = 3.21NSTVASLPAMTISSRR249 pKa = 11.84IRR251 pKa = 11.84FWTKK255 pKa = 9.93FSMLKK260 pKa = 10.47PLDD263 pKa = 3.78TDD265 pKa = 3.79TLQDD269 pKa = 2.93IMTT272 pKa = 4.63
Molecular weight: 30.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629 |
272 |
357 |
314.5 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.175 ± 1.86 | 1.59 ± 0.519 |
4.61 ± 0.103 | 5.246 ± 2.295 |
3.498 ± 0.115 | 5.405 ± 1.498 |
2.226 ± 0.459 | 6.041 ± 0.767 |
4.928 ± 0.803 | 6.2 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.293 ± 0.598 | 4.134 ± 0.393 |
5.246 ± 0.731 | 3.498 ± 0.338 |
8.744 ± 0.719 | 5.723 ± 0.99 |
7.79 ± 1.522 | 4.134 ± 0.839 |
2.544 ± 0.018 | 3.975 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
