
Diabrotica virgifera virgifera (western corn rootworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Coleoptera; Polyphaga; Cucujiformia; Chrysomeloidea; Chrysomelidae; Galerucinae;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
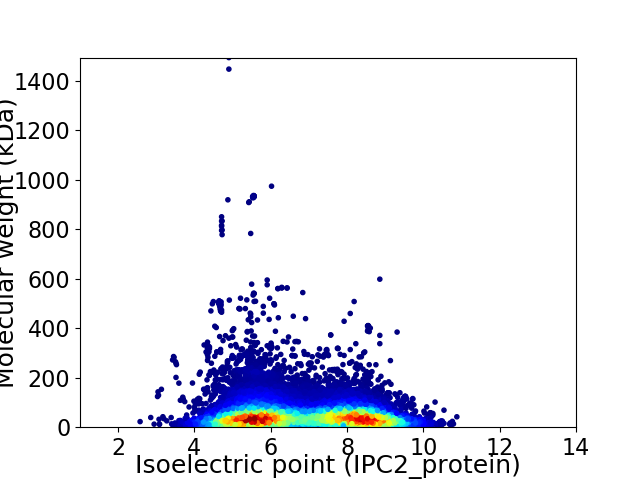
Virtual 2D-PAGE plot for 25502 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
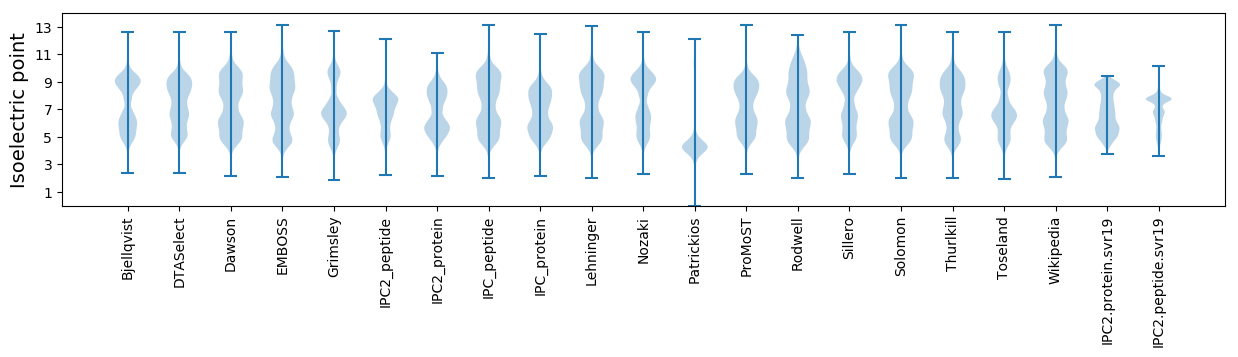
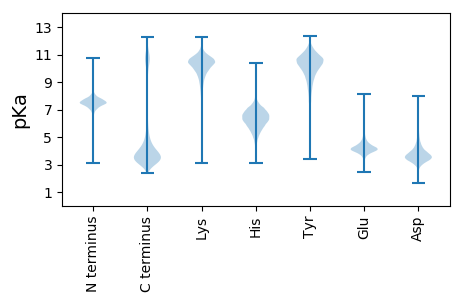
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P7FUQ4|A0A6P7FUQ4_DIAVI uncharacterized protein LOC114331184 OS=Diabrotica virgifera virgifera OX=50390 GN=LOC114331184 PE=4 SV=1
MM1 pKa = 7.55EE2 pKa = 4.93GFIIFLIVATVIGVSALFIKK22 pKa = 9.66TCIKK26 pKa = 10.2CFSVKK31 pKa = 10.15KK32 pKa = 10.19PRR34 pKa = 11.84QSQNNNFVSDD44 pKa = 3.54NYY46 pKa = 10.9DD47 pKa = 3.34YY48 pKa = 11.45EE49 pKa = 6.14DD50 pKa = 3.78GVIFTSEE57 pKa = 4.67LYY59 pKa = 9.3GTDD62 pKa = 3.66YY63 pKa = 11.47NVDD66 pKa = 3.42SVNEE70 pKa = 4.06VTHH73 pKa = 5.82IHH75 pKa = 6.34HH76 pKa = 7.14FEE78 pKa = 3.93TSDD81 pKa = 4.97NIDD84 pKa = 3.64TTPGDD89 pKa = 3.61TDD91 pKa = 3.42NTGSGVASGLAGGFAEE107 pKa = 4.63TFTSSFADD115 pKa = 3.59PGEE118 pKa = 4.13SSTYY122 pKa = 10.58DD123 pKa = 3.14SGGGGFDD130 pKa = 3.31SGGGGDD136 pKa = 3.5TGGGGDD142 pKa = 3.6TGGGGDD148 pKa = 3.37TGGSYY153 pKa = 10.87DD154 pKa = 3.4
MM1 pKa = 7.55EE2 pKa = 4.93GFIIFLIVATVIGVSALFIKK22 pKa = 9.66TCIKK26 pKa = 10.2CFSVKK31 pKa = 10.15KK32 pKa = 10.19PRR34 pKa = 11.84QSQNNNFVSDD44 pKa = 3.54NYY46 pKa = 10.9DD47 pKa = 3.34YY48 pKa = 11.45EE49 pKa = 6.14DD50 pKa = 3.78GVIFTSEE57 pKa = 4.67LYY59 pKa = 9.3GTDD62 pKa = 3.66YY63 pKa = 11.47NVDD66 pKa = 3.42SVNEE70 pKa = 4.06VTHH73 pKa = 5.82IHH75 pKa = 6.34HH76 pKa = 7.14FEE78 pKa = 3.93TSDD81 pKa = 4.97NIDD84 pKa = 3.64TTPGDD89 pKa = 3.61TDD91 pKa = 3.42NTGSGVASGLAGGFAEE107 pKa = 4.63TFTSSFADD115 pKa = 3.59PGEE118 pKa = 4.13SSTYY122 pKa = 10.58DD123 pKa = 3.14SGGGGFDD130 pKa = 3.31SGGGGDD136 pKa = 3.5TGGGGDD142 pKa = 3.6TGGGGDD148 pKa = 3.37TGGSYY153 pKa = 10.87DD154 pKa = 3.4
Molecular weight: 15.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P7GIY1|A0A6P7GIY1_DIAVI zinc finger protein 501-like OS=Diabrotica virgifera virgifera OX=50390 GN=LOC114343270 PE=4 SV=1
VV1 pKa = 7.14KK2 pKa = 10.52NDD4 pKa = 3.64LNDD7 pKa = 3.92VILSNSEE14 pKa = 3.74DD15 pKa = 3.47GKK17 pKa = 11.02ILFQSLIDD25 pKa = 4.13PKK27 pKa = 10.21LTEE30 pKa = 4.99PPPPPPPTDD39 pKa = 3.33PLGPPEE45 pKa = 4.82PPSDD49 pKa = 3.89VPEE52 pKa = 4.57DD53 pKa = 3.83LSNKK57 pKa = 9.5RR58 pKa = 11.84PRR60 pKa = 11.84PPPKK64 pKa = 9.87ALSSKK69 pKa = 10.24RR70 pKa = 11.84PRR72 pKa = 11.84PPPSKK77 pKa = 10.32RR78 pKa = 11.84PRR80 pKa = 11.84TTSPPPSRR88 pKa = 11.84RR89 pKa = 11.84PKK91 pKa = 10.5PPGPPPTRR99 pKa = 11.84RR100 pKa = 11.84PRR102 pKa = 11.84PPGPPPSRR110 pKa = 11.84RR111 pKa = 11.84PRR113 pKa = 11.84PPGPPPSRR121 pKa = 11.84RR122 pKa = 11.84PRR124 pKa = 11.84PPGPPPSRR132 pKa = 11.84RR133 pKa = 11.84PRR135 pKa = 11.84PPGPPPSRR143 pKa = 11.84RR144 pKa = 11.84PRR146 pKa = 11.84PPGPPPSRR154 pKa = 11.84RR155 pKa = 11.84PRR157 pKa = 11.84PPGPPPSRR165 pKa = 11.84RR166 pKa = 11.84PRR168 pKa = 11.84PPGPPPSRR176 pKa = 11.84RR177 pKa = 11.84PRR179 pKa = 11.84PPGPPPSRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84PPGPPPSRR198 pKa = 11.84RR199 pKa = 11.84PRR201 pKa = 11.84PPGPPPSRR209 pKa = 11.84RR210 pKa = 11.84PRR212 pKa = 11.84PPGPPPSRR220 pKa = 11.84RR221 pKa = 11.84PRR223 pKa = 11.84PPGPPPSRR231 pKa = 11.84RR232 pKa = 11.84PRR234 pKa = 11.84PPGPPPSRR242 pKa = 11.84RR243 pKa = 11.84PRR245 pKa = 11.84PPGPPPSRR253 pKa = 11.84RR254 pKa = 11.84PRR256 pKa = 11.84PPGPPPSRR264 pKa = 11.84RR265 pKa = 11.84PRR267 pKa = 11.84PPGPPPSRR275 pKa = 11.84RR276 pKa = 11.84PRR278 pKa = 11.84PPGPPPSRR286 pKa = 11.84RR287 pKa = 11.84PRR289 pKa = 11.84PPGPPPSRR297 pKa = 11.84RR298 pKa = 11.84PRR300 pKa = 11.84PPGPPPSRR308 pKa = 11.84RR309 pKa = 11.84PP310 pKa = 3.28
VV1 pKa = 7.14KK2 pKa = 10.52NDD4 pKa = 3.64LNDD7 pKa = 3.92VILSNSEE14 pKa = 3.74DD15 pKa = 3.47GKK17 pKa = 11.02ILFQSLIDD25 pKa = 4.13PKK27 pKa = 10.21LTEE30 pKa = 4.99PPPPPPPTDD39 pKa = 3.33PLGPPEE45 pKa = 4.82PPSDD49 pKa = 3.89VPEE52 pKa = 4.57DD53 pKa = 3.83LSNKK57 pKa = 9.5RR58 pKa = 11.84PRR60 pKa = 11.84PPPKK64 pKa = 9.87ALSSKK69 pKa = 10.24RR70 pKa = 11.84PRR72 pKa = 11.84PPPSKK77 pKa = 10.32RR78 pKa = 11.84PRR80 pKa = 11.84TTSPPPSRR88 pKa = 11.84RR89 pKa = 11.84PKK91 pKa = 10.5PPGPPPTRR99 pKa = 11.84RR100 pKa = 11.84PRR102 pKa = 11.84PPGPPPSRR110 pKa = 11.84RR111 pKa = 11.84PRR113 pKa = 11.84PPGPPPSRR121 pKa = 11.84RR122 pKa = 11.84PRR124 pKa = 11.84PPGPPPSRR132 pKa = 11.84RR133 pKa = 11.84PRR135 pKa = 11.84PPGPPPSRR143 pKa = 11.84RR144 pKa = 11.84PRR146 pKa = 11.84PPGPPPSRR154 pKa = 11.84RR155 pKa = 11.84PRR157 pKa = 11.84PPGPPPSRR165 pKa = 11.84RR166 pKa = 11.84PRR168 pKa = 11.84PPGPPPSRR176 pKa = 11.84RR177 pKa = 11.84PRR179 pKa = 11.84PPGPPPSRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84PPGPPPSRR198 pKa = 11.84RR199 pKa = 11.84PRR201 pKa = 11.84PPGPPPSRR209 pKa = 11.84RR210 pKa = 11.84PRR212 pKa = 11.84PPGPPPSRR220 pKa = 11.84RR221 pKa = 11.84PRR223 pKa = 11.84PPGPPPSRR231 pKa = 11.84RR232 pKa = 11.84PRR234 pKa = 11.84PPGPPPSRR242 pKa = 11.84RR243 pKa = 11.84PRR245 pKa = 11.84PPGPPPSRR253 pKa = 11.84RR254 pKa = 11.84PRR256 pKa = 11.84PPGPPPSRR264 pKa = 11.84RR265 pKa = 11.84PRR267 pKa = 11.84PPGPPPSRR275 pKa = 11.84RR276 pKa = 11.84PRR278 pKa = 11.84PPGPPPSRR286 pKa = 11.84RR287 pKa = 11.84PRR289 pKa = 11.84PPGPPPSRR297 pKa = 11.84RR298 pKa = 11.84PRR300 pKa = 11.84PPGPPPSRR308 pKa = 11.84RR309 pKa = 11.84PP310 pKa = 3.28
Molecular weight: 33.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12442332 |
30 |
19839 |
487.9 |
55.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.294 ± 0.017 | 2.167 ± 0.025 |
5.498 ± 0.016 | 7.113 ± 0.023 |
3.978 ± 0.014 | 5.116 ± 0.023 |
2.513 ± 0.011 | 6.17 ± 0.013 |
7.647 ± 0.025 | 8.747 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.14 ± 0.01 | 5.555 ± 0.019 |
4.875 ± 0.019 | 4.251 ± 0.019 |
4.72 ± 0.017 | 7.905 ± 0.021 |
6.073 ± 0.027 | 5.998 ± 0.015 |
0.958 ± 0.006 | 3.276 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
