
Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Citrobacter; Citrobacter koseri
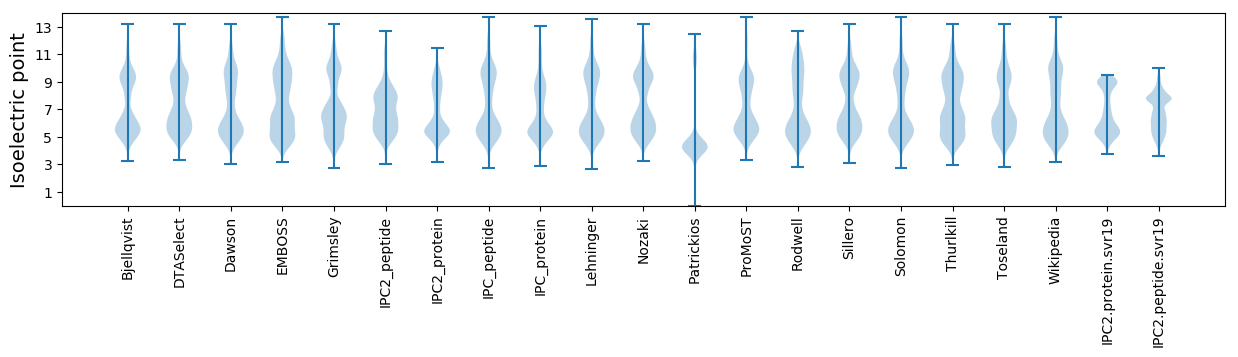
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
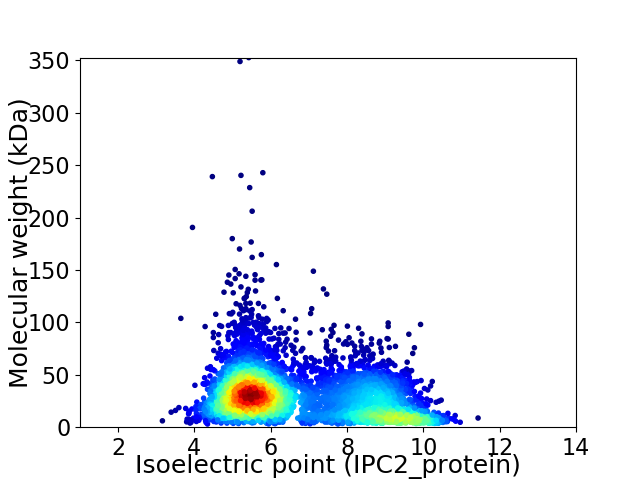
Virtual 2D-PAGE plot for 5019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8AKA3|A8AKA3_CITK8 ABC transmembrane type-1 domain-containing protein OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=CKO_02810 PE=3 SV=1
MM1 pKa = 7.37HH2 pKa = 6.88SWKK5 pKa = 10.29KK6 pKa = 10.49KK7 pKa = 9.88LVVSQLALACTLAITSQANASTDD30 pKa = 3.14ISGTTYY36 pKa = 8.15TTFNHH41 pKa = 6.08YY42 pKa = 10.94NDD44 pKa = 3.27ATYY47 pKa = 11.45ADD49 pKa = 3.54GVYY52 pKa = 10.55YY53 pKa = 10.6DD54 pKa = 5.91GYY56 pKa = 11.26VGWNNYY62 pKa = 7.95ATDD65 pKa = 3.4SVYY68 pKa = 11.18NGDD71 pKa = 3.99IYY73 pKa = 10.88PVINNATVNGVISTYY88 pKa = 11.01YY89 pKa = 10.75LDD91 pKa = 5.97DD92 pKa = 4.34GLSTNTNSNSLTIKK106 pKa = 10.46NSTIHH111 pKa = 6.74GMITSEE117 pKa = 5.7CMTTDD122 pKa = 3.19CVGRR126 pKa = 11.84DD127 pKa = 3.38ATGYY131 pKa = 10.62VYY133 pKa = 10.81DD134 pKa = 4.51RR135 pKa = 11.84LALTVDD141 pKa = 3.46NSTIDD146 pKa = 3.71DD147 pKa = 3.99NYY149 pKa = 11.03EE150 pKa = 3.75HH151 pKa = 6.27YY152 pKa = 9.43TYY154 pKa = 11.16NGTYY158 pKa = 9.91TDD160 pKa = 3.91GTADD164 pKa = 3.28THH166 pKa = 7.24VVDD169 pKa = 5.12VYY171 pKa = 11.89NLGTAITLDD180 pKa = 3.51QEE182 pKa = 3.92VDD184 pKa = 3.74LVIKK188 pKa = 10.62NNSHH192 pKa = 5.43VAGITLTQGYY202 pKa = 8.14EE203 pKa = 3.79WEE205 pKa = 5.73DD206 pKa = 3.2IDD208 pKa = 6.2DD209 pKa = 4.15NTVSTGVNSAEE220 pKa = 4.11VFNNTVTVTDD230 pKa = 3.63STVTSGSWSDD240 pKa = 3.49EE241 pKa = 3.9GTSGWFGNTNNASDD255 pKa = 4.14YY256 pKa = 10.24SDD258 pKa = 5.06SYY260 pKa = 10.42TADD263 pKa = 3.4DD264 pKa = 3.7VAIAAIAHH272 pKa = 6.19PNADD276 pKa = 3.04NAMQTTVNLSNSTLMGDD293 pKa = 3.78VVFSSNFDD301 pKa = 3.48EE302 pKa = 5.33NFFPNGADD310 pKa = 3.41TYY312 pKa = 11.03RR313 pKa = 11.84DD314 pKa = 3.14TDD316 pKa = 3.75ADD318 pKa = 4.56LDD320 pKa = 4.29TNGWDD325 pKa = 3.47GTDD328 pKa = 3.71RR329 pKa = 11.84MDD331 pKa = 3.14VTLNNGSKK339 pKa = 9.47WVGAAMSVHH348 pKa = 5.8QVDD351 pKa = 3.33SDD353 pKa = 3.81GDD355 pKa = 3.83GVYY358 pKa = 10.9DD359 pKa = 4.0SFAAGTDD366 pKa = 3.41ATATLIDD373 pKa = 3.52IAANSLWPSSTYY385 pKa = 11.0GIDD388 pKa = 3.95NNDD391 pKa = 3.02TAYY394 pKa = 10.63GEE396 pKa = 4.72DD397 pKa = 3.26GHH399 pKa = 7.61VVGNEE404 pKa = 4.1VYY406 pKa = 10.48QSGLFNVTLNGGSQWDD422 pKa = 3.84TTKK425 pKa = 10.85TSLIDD430 pKa = 3.4TLSINSGSVVNVADD444 pKa = 3.84SDD446 pKa = 4.34LVSDD450 pKa = 5.06SISLTGGAALNINEE464 pKa = 5.08DD465 pKa = 3.42GHH467 pKa = 6.03VATDD471 pKa = 3.47EE472 pKa = 4.06LTINNSTVTIADD484 pKa = 3.86DD485 pKa = 4.15VSAGWGVYY493 pKa = 9.5DD494 pKa = 4.01AALYY498 pKa = 11.08ANTINVTNNGVLDD511 pKa = 3.97VGNSTAYY518 pKa = 10.41ALQADD523 pKa = 4.43TLNLTSYY530 pKa = 10.6TDD532 pKa = 3.38ANGNVNAGVFNVHH545 pKa = 6.3SNSFVLDD552 pKa = 3.52ADD554 pKa = 3.95LTNDD558 pKa = 3.33RR559 pKa = 11.84TNDD562 pKa = 3.17ITKK565 pKa = 10.28SNYY568 pKa = 9.27GYY570 pKa = 11.19GVIAMNSDD578 pKa = 2.85GHH580 pKa = 5.83LTINGNGDD588 pKa = 3.08AWTGDD593 pKa = 3.25QSEE596 pKa = 4.42VDD598 pKa = 3.37NAGDD602 pKa = 3.43NVAAATGNYY611 pKa = 8.97KK612 pKa = 10.02VRR614 pKa = 11.84IDD616 pKa = 3.45NATGEE621 pKa = 4.47GSVADD626 pKa = 3.94YY627 pKa = 10.58KK628 pKa = 11.15GKK630 pKa = 10.29EE631 pKa = 3.97LIYY634 pKa = 11.34VNDD637 pKa = 4.19KK638 pKa = 10.67NSKK641 pKa = 8.67ATFSAANKK649 pKa = 9.86ADD651 pKa = 3.72LGAYY655 pKa = 7.63TYY657 pKa = 10.27QAQQEE662 pKa = 4.68GNTVVMQQMEE672 pKa = 4.41LTDD675 pKa = 4.11YY676 pKa = 11.84ANMALSIPSANTNIWNLEE694 pKa = 3.6QDD696 pKa = 3.86TVGTRR701 pKa = 11.84LTNSRR706 pKa = 11.84HH707 pKa = 4.84GLADD711 pKa = 3.5NGGAWVSYY719 pKa = 10.31FGGSFDD725 pKa = 5.18GDD727 pKa = 3.32NGTINYY733 pKa = 8.82DD734 pKa = 3.13QDD736 pKa = 3.61VNGIMVGVDD745 pKa = 2.97TKK747 pKa = 11.02IDD749 pKa = 3.49GNNAKK754 pKa = 9.7WIVGAAAGFAKK765 pKa = 10.6GDD767 pKa = 3.43MSDD770 pKa = 2.99RR771 pKa = 11.84TGQVDD776 pKa = 3.54QDD778 pKa = 3.84SQSAYY783 pKa = 8.65IYY785 pKa = 10.6SSARR789 pKa = 11.84FANNVFVDD797 pKa = 4.71GSLSYY802 pKa = 11.41SHH804 pKa = 7.31FNNDD808 pKa = 3.29LSANMSNGQYY818 pKa = 10.78VDD820 pKa = 3.92GNTSADD826 pKa = 2.84AWGFGLKK833 pKa = 10.32LGYY836 pKa = 9.47DD837 pKa = 3.67WKK839 pKa = 11.09LGDD842 pKa = 3.96AGYY845 pKa = 8.07VTPYY849 pKa = 10.65GSVSGLFQSGDD860 pKa = 3.79DD861 pKa = 3.89YY862 pKa = 11.83QLSNDD867 pKa = 3.96MKK869 pKa = 11.23VDD871 pKa = 3.62GQSYY875 pKa = 10.64DD876 pKa = 3.02SMRR879 pKa = 11.84YY880 pKa = 8.94EE881 pKa = 4.34LGVDD885 pKa = 3.21AGYY888 pKa = 8.53TFTYY892 pKa = 10.6SEE894 pKa = 4.47DD895 pKa = 3.66QALTPYY901 pKa = 10.37FKK903 pKa = 10.64LAYY906 pKa = 10.48VYY908 pKa = 11.28DD909 pKa = 4.46DD910 pKa = 4.6ANNDD914 pKa = 3.18ADD916 pKa = 4.23VNGDD920 pKa = 3.84SIDD923 pKa = 3.7NGVEE927 pKa = 3.97GSAVRR932 pKa = 11.84VGLGTQFSFTKK943 pKa = 10.49NFSAYY948 pKa = 9.3TDD950 pKa = 3.45ANYY953 pKa = 10.81LGGGDD958 pKa = 4.6VDD960 pKa = 4.32QDD962 pKa = 2.67WSANVGVKK970 pKa = 7.43YY971 pKa = 8.84TWW973 pKa = 3.09
MM1 pKa = 7.37HH2 pKa = 6.88SWKK5 pKa = 10.29KK6 pKa = 10.49KK7 pKa = 9.88LVVSQLALACTLAITSQANASTDD30 pKa = 3.14ISGTTYY36 pKa = 8.15TTFNHH41 pKa = 6.08YY42 pKa = 10.94NDD44 pKa = 3.27ATYY47 pKa = 11.45ADD49 pKa = 3.54GVYY52 pKa = 10.55YY53 pKa = 10.6DD54 pKa = 5.91GYY56 pKa = 11.26VGWNNYY62 pKa = 7.95ATDD65 pKa = 3.4SVYY68 pKa = 11.18NGDD71 pKa = 3.99IYY73 pKa = 10.88PVINNATVNGVISTYY88 pKa = 11.01YY89 pKa = 10.75LDD91 pKa = 5.97DD92 pKa = 4.34GLSTNTNSNSLTIKK106 pKa = 10.46NSTIHH111 pKa = 6.74GMITSEE117 pKa = 5.7CMTTDD122 pKa = 3.19CVGRR126 pKa = 11.84DD127 pKa = 3.38ATGYY131 pKa = 10.62VYY133 pKa = 10.81DD134 pKa = 4.51RR135 pKa = 11.84LALTVDD141 pKa = 3.46NSTIDD146 pKa = 3.71DD147 pKa = 3.99NYY149 pKa = 11.03EE150 pKa = 3.75HH151 pKa = 6.27YY152 pKa = 9.43TYY154 pKa = 11.16NGTYY158 pKa = 9.91TDD160 pKa = 3.91GTADD164 pKa = 3.28THH166 pKa = 7.24VVDD169 pKa = 5.12VYY171 pKa = 11.89NLGTAITLDD180 pKa = 3.51QEE182 pKa = 3.92VDD184 pKa = 3.74LVIKK188 pKa = 10.62NNSHH192 pKa = 5.43VAGITLTQGYY202 pKa = 8.14EE203 pKa = 3.79WEE205 pKa = 5.73DD206 pKa = 3.2IDD208 pKa = 6.2DD209 pKa = 4.15NTVSTGVNSAEE220 pKa = 4.11VFNNTVTVTDD230 pKa = 3.63STVTSGSWSDD240 pKa = 3.49EE241 pKa = 3.9GTSGWFGNTNNASDD255 pKa = 4.14YY256 pKa = 10.24SDD258 pKa = 5.06SYY260 pKa = 10.42TADD263 pKa = 3.4DD264 pKa = 3.7VAIAAIAHH272 pKa = 6.19PNADD276 pKa = 3.04NAMQTTVNLSNSTLMGDD293 pKa = 3.78VVFSSNFDD301 pKa = 3.48EE302 pKa = 5.33NFFPNGADD310 pKa = 3.41TYY312 pKa = 11.03RR313 pKa = 11.84DD314 pKa = 3.14TDD316 pKa = 3.75ADD318 pKa = 4.56LDD320 pKa = 4.29TNGWDD325 pKa = 3.47GTDD328 pKa = 3.71RR329 pKa = 11.84MDD331 pKa = 3.14VTLNNGSKK339 pKa = 9.47WVGAAMSVHH348 pKa = 5.8QVDD351 pKa = 3.33SDD353 pKa = 3.81GDD355 pKa = 3.83GVYY358 pKa = 10.9DD359 pKa = 4.0SFAAGTDD366 pKa = 3.41ATATLIDD373 pKa = 3.52IAANSLWPSSTYY385 pKa = 11.0GIDD388 pKa = 3.95NNDD391 pKa = 3.02TAYY394 pKa = 10.63GEE396 pKa = 4.72DD397 pKa = 3.26GHH399 pKa = 7.61VVGNEE404 pKa = 4.1VYY406 pKa = 10.48QSGLFNVTLNGGSQWDD422 pKa = 3.84TTKK425 pKa = 10.85TSLIDD430 pKa = 3.4TLSINSGSVVNVADD444 pKa = 3.84SDD446 pKa = 4.34LVSDD450 pKa = 5.06SISLTGGAALNINEE464 pKa = 5.08DD465 pKa = 3.42GHH467 pKa = 6.03VATDD471 pKa = 3.47EE472 pKa = 4.06LTINNSTVTIADD484 pKa = 3.86DD485 pKa = 4.15VSAGWGVYY493 pKa = 9.5DD494 pKa = 4.01AALYY498 pKa = 11.08ANTINVTNNGVLDD511 pKa = 3.97VGNSTAYY518 pKa = 10.41ALQADD523 pKa = 4.43TLNLTSYY530 pKa = 10.6TDD532 pKa = 3.38ANGNVNAGVFNVHH545 pKa = 6.3SNSFVLDD552 pKa = 3.52ADD554 pKa = 3.95LTNDD558 pKa = 3.33RR559 pKa = 11.84TNDD562 pKa = 3.17ITKK565 pKa = 10.28SNYY568 pKa = 9.27GYY570 pKa = 11.19GVIAMNSDD578 pKa = 2.85GHH580 pKa = 5.83LTINGNGDD588 pKa = 3.08AWTGDD593 pKa = 3.25QSEE596 pKa = 4.42VDD598 pKa = 3.37NAGDD602 pKa = 3.43NVAAATGNYY611 pKa = 8.97KK612 pKa = 10.02VRR614 pKa = 11.84IDD616 pKa = 3.45NATGEE621 pKa = 4.47GSVADD626 pKa = 3.94YY627 pKa = 10.58KK628 pKa = 11.15GKK630 pKa = 10.29EE631 pKa = 3.97LIYY634 pKa = 11.34VNDD637 pKa = 4.19KK638 pKa = 10.67NSKK641 pKa = 8.67ATFSAANKK649 pKa = 9.86ADD651 pKa = 3.72LGAYY655 pKa = 7.63TYY657 pKa = 10.27QAQQEE662 pKa = 4.68GNTVVMQQMEE672 pKa = 4.41LTDD675 pKa = 4.11YY676 pKa = 11.84ANMALSIPSANTNIWNLEE694 pKa = 3.6QDD696 pKa = 3.86TVGTRR701 pKa = 11.84LTNSRR706 pKa = 11.84HH707 pKa = 4.84GLADD711 pKa = 3.5NGGAWVSYY719 pKa = 10.31FGGSFDD725 pKa = 5.18GDD727 pKa = 3.32NGTINYY733 pKa = 8.82DD734 pKa = 3.13QDD736 pKa = 3.61VNGIMVGVDD745 pKa = 2.97TKK747 pKa = 11.02IDD749 pKa = 3.49GNNAKK754 pKa = 9.7WIVGAAAGFAKK765 pKa = 10.6GDD767 pKa = 3.43MSDD770 pKa = 2.99RR771 pKa = 11.84TGQVDD776 pKa = 3.54QDD778 pKa = 3.84SQSAYY783 pKa = 8.65IYY785 pKa = 10.6SSARR789 pKa = 11.84FANNVFVDD797 pKa = 4.71GSLSYY802 pKa = 11.41SHH804 pKa = 7.31FNNDD808 pKa = 3.29LSANMSNGQYY818 pKa = 10.78VDD820 pKa = 3.92GNTSADD826 pKa = 2.84AWGFGLKK833 pKa = 10.32LGYY836 pKa = 9.47DD837 pKa = 3.67WKK839 pKa = 11.09LGDD842 pKa = 3.96AGYY845 pKa = 8.07VTPYY849 pKa = 10.65GSVSGLFQSGDD860 pKa = 3.79DD861 pKa = 3.89YY862 pKa = 11.83QLSNDD867 pKa = 3.96MKK869 pKa = 11.23VDD871 pKa = 3.62GQSYY875 pKa = 10.64DD876 pKa = 3.02SMRR879 pKa = 11.84YY880 pKa = 8.94EE881 pKa = 4.34LGVDD885 pKa = 3.21AGYY888 pKa = 8.53TFTYY892 pKa = 10.6SEE894 pKa = 4.47DD895 pKa = 3.66QALTPYY901 pKa = 10.37FKK903 pKa = 10.64LAYY906 pKa = 10.48VYY908 pKa = 11.28DD909 pKa = 4.46DD910 pKa = 4.6ANNDD914 pKa = 3.18ADD916 pKa = 4.23VNGDD920 pKa = 3.84SIDD923 pKa = 3.7NGVEE927 pKa = 3.97GSAVRR932 pKa = 11.84VGLGTQFSFTKK943 pKa = 10.49NFSAYY948 pKa = 9.3TDD950 pKa = 3.45ANYY953 pKa = 10.81LGGGDD958 pKa = 4.6VDD960 pKa = 4.32QDD962 pKa = 2.67WSANVGVKK970 pKa = 7.43YY971 pKa = 8.84TWW973 pKa = 3.09
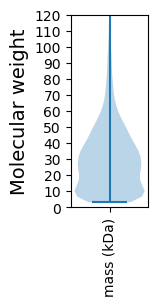
Molecular weight: 103.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8AMV3|A8AMV3_CITK8 Uncharacterized protein OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=CKO_03740 PE=4 SV=1
MM1 pKa = 6.45VARR4 pKa = 11.84WRR6 pKa = 11.84KK7 pKa = 9.95LIGPTISPHH16 pKa = 6.17RR17 pKa = 11.84STVGRR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84SRR27 pKa = 11.84HH28 pKa = 4.65PALPAPHH35 pKa = 7.43RR36 pKa = 11.84STVGRR41 pKa = 11.84IRR43 pKa = 11.84RR44 pKa = 11.84SHH46 pKa = 6.56HH47 pKa = 6.09PALPAPHH54 pKa = 7.3RR55 pKa = 11.84NTVGRR60 pKa = 11.84IRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84HH66 pKa = 4.9PALLPGIPARR76 pKa = 11.84FF77 pKa = 3.37
MM1 pKa = 6.45VARR4 pKa = 11.84WRR6 pKa = 11.84KK7 pKa = 9.95LIGPTISPHH16 pKa = 6.17RR17 pKa = 11.84STVGRR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84SRR27 pKa = 11.84HH28 pKa = 4.65PALPAPHH35 pKa = 7.43RR36 pKa = 11.84STVGRR41 pKa = 11.84IRR43 pKa = 11.84RR44 pKa = 11.84SHH46 pKa = 6.56HH47 pKa = 6.09PALPAPHH54 pKa = 7.3RR55 pKa = 11.84NTVGRR60 pKa = 11.84IRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84HH66 pKa = 4.9PALLPGIPARR76 pKa = 11.84FF77 pKa = 3.37
Molecular weight: 8.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1442289 |
27 |
3208 |
287.4 |
31.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.746 ± 0.048 | 1.176 ± 0.015 |
5.149 ± 0.026 | 5.506 ± 0.033 |
3.963 ± 0.027 | 7.386 ± 0.035 |
2.334 ± 0.017 | 5.862 ± 0.028 |
4.132 ± 0.031 | 10.754 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.811 ± 0.018 | 3.791 ± 0.027 |
4.447 ± 0.031 | 4.477 ± 0.031 |
5.788 ± 0.033 | 5.876 ± 0.026 |
5.446 ± 0.025 | 7.08 ± 0.03 |
1.503 ± 0.015 | 2.775 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
