
Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) (Filobasidiella neoformans)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Cryptococcus; Cryptococcus neoformans species complex; Cryptococcus neoformans; Cryptococcus neoformans var. neoformans
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
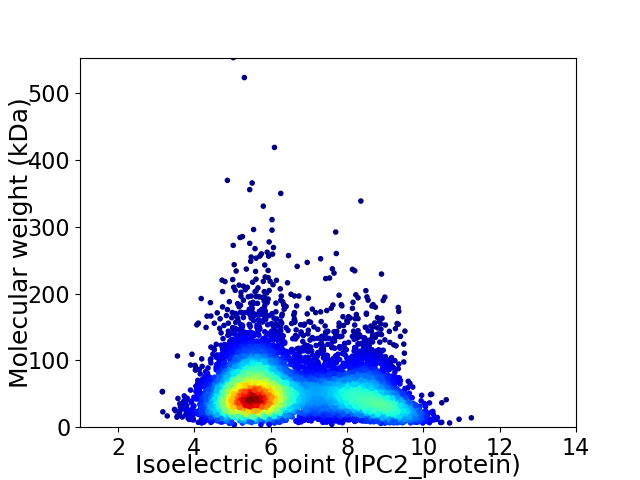
Virtual 2D-PAGE plot for 6746 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2M5N0|A0A0S2M5N0_CRYNJ Uncharacterized protein OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CNF02385 PE=4 SV=1
MM1 pKa = 7.04TLKK4 pKa = 10.16IQRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.77LTAIQKK15 pKa = 7.96MISVKK20 pKa = 7.62QTIPFGLLSCLALLGSIKK38 pKa = 10.76ADD40 pKa = 3.08DD41 pKa = 4.01STNNTSWPDD50 pKa = 3.23WVTNDD55 pKa = 3.36YY56 pKa = 11.43DD57 pKa = 4.33CVIGCLSGFNDD68 pKa = 5.0TITSVPQDD76 pKa = 3.65ALAQQAFDD84 pKa = 4.5CTSSTCTGDD93 pKa = 2.92ATGNYY98 pKa = 8.4YY99 pKa = 8.24QTFYY103 pKa = 11.13YY104 pKa = 9.84IQLFYY109 pKa = 10.54ATGSIYY115 pKa = 10.36EE116 pKa = 4.51SSDD119 pKa = 3.46SAPDD123 pKa = 4.31GYY125 pKa = 11.13KK126 pKa = 10.09HH127 pKa = 6.9AAFTDD132 pKa = 3.93STDD135 pKa = 3.57TTSEE139 pKa = 4.03AAVSGDD145 pKa = 3.03WGDD148 pKa = 4.85DD149 pKa = 3.44DD150 pKa = 5.34TVDD153 pKa = 3.6PTEE156 pKa = 5.77DD157 pKa = 2.97IAASDD162 pKa = 3.89SASATDD168 pKa = 3.48SAEE171 pKa = 4.25TTGSADD177 pKa = 3.52GVDD180 pKa = 3.92GGSATDD186 pKa = 3.63TAALAIDD193 pKa = 3.84TGTEE197 pKa = 3.5NGTVVTGTAADD208 pKa = 3.85STASGSKK215 pKa = 10.19LSSASHH221 pKa = 6.45TSSQSDD227 pKa = 3.22ASTASSSVAADD238 pKa = 3.18ISEE241 pKa = 4.08SGAFKK246 pKa = 10.55TIGGDD251 pKa = 3.16VAQRR255 pKa = 11.84IFGAVGAMVFGGLWIGLL272 pKa = 3.88
MM1 pKa = 7.04TLKK4 pKa = 10.16IQRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.77LTAIQKK15 pKa = 7.96MISVKK20 pKa = 7.62QTIPFGLLSCLALLGSIKK38 pKa = 10.76ADD40 pKa = 3.08DD41 pKa = 4.01STNNTSWPDD50 pKa = 3.23WVTNDD55 pKa = 3.36YY56 pKa = 11.43DD57 pKa = 4.33CVIGCLSGFNDD68 pKa = 5.0TITSVPQDD76 pKa = 3.65ALAQQAFDD84 pKa = 4.5CTSSTCTGDD93 pKa = 2.92ATGNYY98 pKa = 8.4YY99 pKa = 8.24QTFYY103 pKa = 11.13YY104 pKa = 9.84IQLFYY109 pKa = 10.54ATGSIYY115 pKa = 10.36EE116 pKa = 4.51SSDD119 pKa = 3.46SAPDD123 pKa = 4.31GYY125 pKa = 11.13KK126 pKa = 10.09HH127 pKa = 6.9AAFTDD132 pKa = 3.93STDD135 pKa = 3.57TTSEE139 pKa = 4.03AAVSGDD145 pKa = 3.03WGDD148 pKa = 4.85DD149 pKa = 3.44DD150 pKa = 5.34TVDD153 pKa = 3.6PTEE156 pKa = 5.77DD157 pKa = 2.97IAASDD162 pKa = 3.89SASATDD168 pKa = 3.48SAEE171 pKa = 4.25TTGSADD177 pKa = 3.52GVDD180 pKa = 3.92GGSATDD186 pKa = 3.63TAALAIDD193 pKa = 3.84TGTEE197 pKa = 3.5NGTVVTGTAADD208 pKa = 3.85STASGSKK215 pKa = 10.19LSSASHH221 pKa = 6.45TSSQSDD227 pKa = 3.22ASTASSSVAADD238 pKa = 3.18ISEE241 pKa = 4.08SGAFKK246 pKa = 10.55TIGGDD251 pKa = 3.16VAQRR255 pKa = 11.84IFGAVGAMVFGGLWIGLL272 pKa = 3.88
Molecular weight: 27.84 kDa
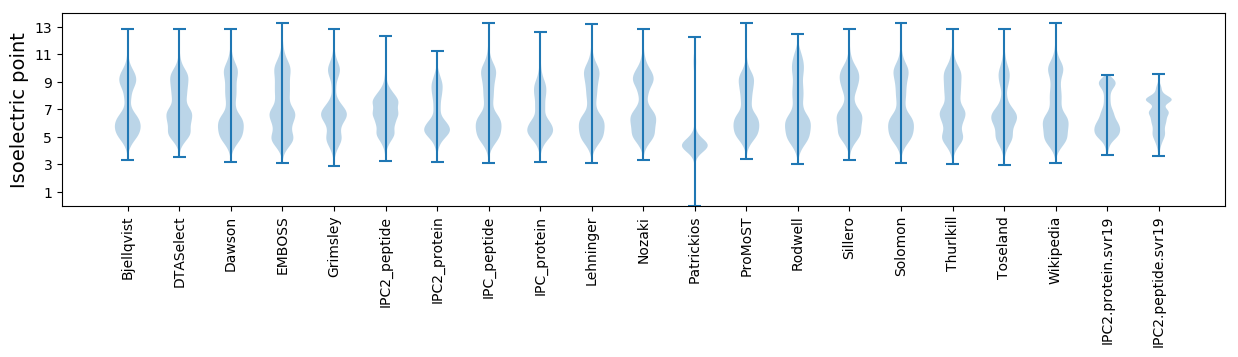
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2LIC9|A0A0S2LIC9_CRYNJ Uncharacterized protein OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CNC02885 PE=4 SV=1
MM1 pKa = 7.45PRR3 pKa = 11.84LARR6 pKa = 11.84VLAVPLPPRR15 pKa = 11.84ARR17 pKa = 11.84LAPLSLPRR25 pKa = 11.84ATLSAPAPAPQSSILSSLRR44 pKa = 11.84TPTSTFVASRR54 pKa = 11.84PMALSSPPIPTHH66 pKa = 6.48ILPSLPAFSPLGALRR81 pKa = 11.84HH82 pKa = 4.92VAMGAFYY89 pKa = 10.66QPSQRR94 pKa = 11.84KK95 pKa = 8.72RR96 pKa = 11.84KK97 pKa = 8.74NKK99 pKa = 9.96HH100 pKa = 5.07GFLSRR105 pKa = 11.84LKK107 pKa = 10.47GGKK110 pKa = 9.1NGRR113 pKa = 11.84KK114 pKa = 6.94TLLRR118 pKa = 11.84RR119 pKa = 11.84LLKK122 pKa = 10.03GRR124 pKa = 11.84KK125 pKa = 8.54FLSHH129 pKa = 6.98
MM1 pKa = 7.45PRR3 pKa = 11.84LARR6 pKa = 11.84VLAVPLPPRR15 pKa = 11.84ARR17 pKa = 11.84LAPLSLPRR25 pKa = 11.84ATLSAPAPAPQSSILSSLRR44 pKa = 11.84TPTSTFVASRR54 pKa = 11.84PMALSSPPIPTHH66 pKa = 6.48ILPSLPAFSPLGALRR81 pKa = 11.84HH82 pKa = 4.92VAMGAFYY89 pKa = 10.66QPSQRR94 pKa = 11.84KK95 pKa = 8.72RR96 pKa = 11.84KK97 pKa = 8.74NKK99 pKa = 9.96HH100 pKa = 5.07GFLSRR105 pKa = 11.84LKK107 pKa = 10.47GGKK110 pKa = 9.1NGRR113 pKa = 11.84KK114 pKa = 6.94TLLRR118 pKa = 11.84RR119 pKa = 11.84LLKK122 pKa = 10.03GRR124 pKa = 11.84KK125 pKa = 8.54FLSHH129 pKa = 6.98
Molecular weight: 14.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3636027 |
37 |
4972 |
539.0 |
59.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.362 ± 0.026 | 1.024 ± 0.01 |
5.376 ± 0.018 | 6.614 ± 0.029 |
3.406 ± 0.018 | 7.095 ± 0.025 |
2.343 ± 0.012 | 4.798 ± 0.019 |
5.19 ± 0.026 | 8.884 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.153 ± 0.01 | 3.405 ± 0.013 |
6.682 ± 0.036 | 3.864 ± 0.021 |
5.98 ± 0.024 | 9.082 ± 0.037 |
5.796 ± 0.019 | 6.036 ± 0.02 |
1.376 ± 0.011 | 2.533 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
