
Carboxylicivirga sp. M1479
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinilabiliaceae; Carboxylicivirga; unclassified Carboxylicivirga
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
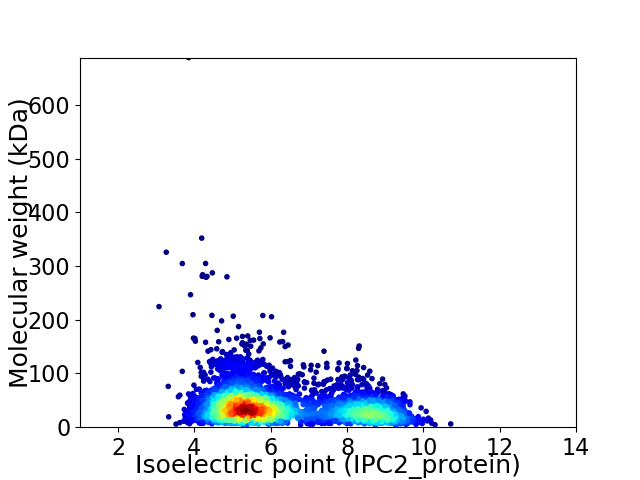
Virtual 2D-PAGE plot for 4095 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
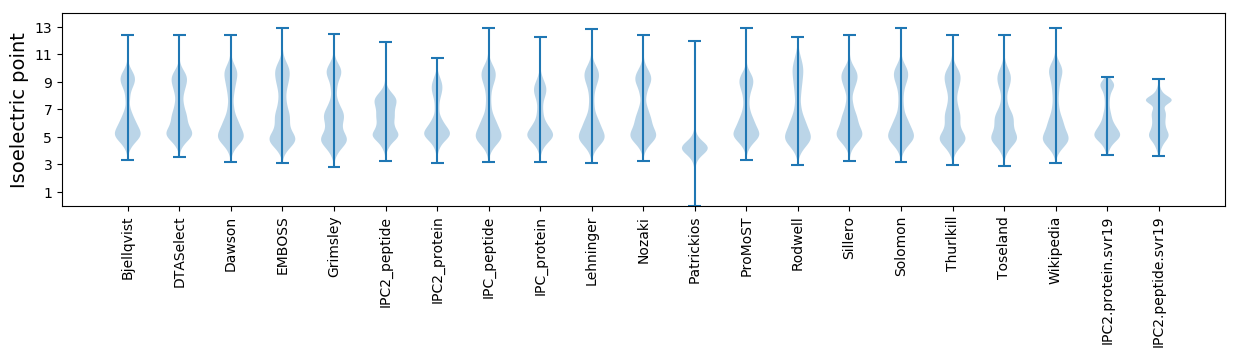
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553FRB2|A0A553FRB2_9BACT Uncharacterized protein OS=Carboxylicivirga sp. M1479 OX=2594476 GN=FNN09_20845 PE=4 SV=1
MM1 pKa = 7.46LLVLVVAFSACKK13 pKa = 10.47DD14 pKa = 4.06DD15 pKa = 6.39DD16 pKa = 5.94DD17 pKa = 7.39DD18 pKa = 5.17DD19 pKa = 6.06APTTGIIKK27 pKa = 8.81GTVTDD32 pKa = 3.8AANNEE37 pKa = 4.24TLSDD41 pKa = 3.64VSIMVFDD48 pKa = 3.68ATTNEE53 pKa = 4.62TIGEE57 pKa = 4.44TIMSGADD64 pKa = 3.05GMYY67 pKa = 10.43SVTLDD72 pKa = 3.24PGTYY76 pKa = 9.17YY77 pKa = 11.18LKK79 pKa = 10.73LFKK82 pKa = 10.18QGYY85 pKa = 8.35NNNPPAGVPPLNISVSEE102 pKa = 4.2GSEE105 pKa = 4.07SVNDD109 pKa = 4.19FKK111 pKa = 11.16MQASEE116 pKa = 4.12VLDD119 pKa = 4.18GGSLTGKK126 pKa = 10.42VMLGSNGVQGAYY138 pKa = 10.67VLVTSGTMGYY148 pKa = 9.85SAVSDD153 pKa = 3.52KK154 pKa = 11.19DD155 pKa = 3.32GHH157 pKa = 5.5FFIYY161 pKa = 10.07NVPAATYY168 pKa = 8.95TLTAYY173 pKa = 9.78KK174 pKa = 9.76INHH177 pKa = 6.53NATPLSVTVTADD189 pKa = 2.98ANTEE193 pKa = 4.23GNVNLTEE200 pKa = 4.4GASTSLSGTLTFLATEE216 pKa = 4.43GKK218 pKa = 9.86DD219 pKa = 3.38VNVNLLHH226 pKa = 6.5PTTNEE231 pKa = 4.46PIPGLVDD238 pKa = 3.16MSEE241 pKa = 3.94NMSYY245 pKa = 10.71SLSGIPDD252 pKa = 3.2GDD254 pKa = 4.05YY255 pKa = 10.8LVKK258 pKa = 10.82ASFEE262 pKa = 4.0NDD264 pKa = 3.31TYY266 pKa = 11.73VVDD269 pKa = 4.53PDD271 pKa = 4.17WIIKK275 pKa = 10.25NGEE278 pKa = 3.96PMLTANGSAVVIDD291 pKa = 4.78FSLTNAVTLNSPTNPMSTTVPIEE314 pKa = 4.02IASTTPTFEE323 pKa = 3.77WTSYY327 pKa = 10.8SSSSDD332 pKa = 3.46YY333 pKa = 11.17VIEE336 pKa = 4.38VSDD339 pKa = 4.22ANGNVIWGGINDD351 pKa = 4.43GVDD354 pKa = 3.42PPLKK358 pKa = 10.55NITIPSSTTSIAYY371 pKa = 10.23NSDD374 pKa = 2.73GMATIPNLEE383 pKa = 3.98YY384 pKa = 10.94DD385 pKa = 3.62VVYY388 pKa = 10.11RR389 pKa = 11.84WKK391 pKa = 10.4IYY393 pKa = 10.05ASKK396 pKa = 11.26NNVQEE401 pKa = 3.85PSGWNLISVSEE412 pKa = 4.09DD413 pKa = 3.12QMGLFIVKK421 pKa = 8.96EE422 pKa = 4.08QQQ424 pKa = 2.75
MM1 pKa = 7.46LLVLVVAFSACKK13 pKa = 10.47DD14 pKa = 4.06DD15 pKa = 6.39DD16 pKa = 5.94DD17 pKa = 7.39DD18 pKa = 5.17DD19 pKa = 6.06APTTGIIKK27 pKa = 8.81GTVTDD32 pKa = 3.8AANNEE37 pKa = 4.24TLSDD41 pKa = 3.64VSIMVFDD48 pKa = 3.68ATTNEE53 pKa = 4.62TIGEE57 pKa = 4.44TIMSGADD64 pKa = 3.05GMYY67 pKa = 10.43SVTLDD72 pKa = 3.24PGTYY76 pKa = 9.17YY77 pKa = 11.18LKK79 pKa = 10.73LFKK82 pKa = 10.18QGYY85 pKa = 8.35NNNPPAGVPPLNISVSEE102 pKa = 4.2GSEE105 pKa = 4.07SVNDD109 pKa = 4.19FKK111 pKa = 11.16MQASEE116 pKa = 4.12VLDD119 pKa = 4.18GGSLTGKK126 pKa = 10.42VMLGSNGVQGAYY138 pKa = 10.67VLVTSGTMGYY148 pKa = 9.85SAVSDD153 pKa = 3.52KK154 pKa = 11.19DD155 pKa = 3.32GHH157 pKa = 5.5FFIYY161 pKa = 10.07NVPAATYY168 pKa = 8.95TLTAYY173 pKa = 9.78KK174 pKa = 9.76INHH177 pKa = 6.53NATPLSVTVTADD189 pKa = 2.98ANTEE193 pKa = 4.23GNVNLTEE200 pKa = 4.4GASTSLSGTLTFLATEE216 pKa = 4.43GKK218 pKa = 9.86DD219 pKa = 3.38VNVNLLHH226 pKa = 6.5PTTNEE231 pKa = 4.46PIPGLVDD238 pKa = 3.16MSEE241 pKa = 3.94NMSYY245 pKa = 10.71SLSGIPDD252 pKa = 3.2GDD254 pKa = 4.05YY255 pKa = 10.8LVKK258 pKa = 10.82ASFEE262 pKa = 4.0NDD264 pKa = 3.31TYY266 pKa = 11.73VVDD269 pKa = 4.53PDD271 pKa = 4.17WIIKK275 pKa = 10.25NGEE278 pKa = 3.96PMLTANGSAVVIDD291 pKa = 4.78FSLTNAVTLNSPTNPMSTTVPIEE314 pKa = 4.02IASTTPTFEE323 pKa = 3.77WTSYY327 pKa = 10.8SSSSDD332 pKa = 3.46YY333 pKa = 11.17VIEE336 pKa = 4.38VSDD339 pKa = 4.22ANGNVIWGGINDD351 pKa = 4.43GVDD354 pKa = 3.42PPLKK358 pKa = 10.55NITIPSSTTSIAYY371 pKa = 10.23NSDD374 pKa = 2.73GMATIPNLEE383 pKa = 3.98YY384 pKa = 10.94DD385 pKa = 3.62VVYY388 pKa = 10.11RR389 pKa = 11.84WKK391 pKa = 10.4IYY393 pKa = 10.05ASKK396 pKa = 11.26NNVQEE401 pKa = 3.85PSGWNLISVSEE412 pKa = 4.09DD413 pKa = 3.12QMGLFIVKK421 pKa = 8.96EE422 pKa = 4.08QQQ424 pKa = 2.75
Molecular weight: 45.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553GPZ8|A0A553GPZ8_9BACT Uncharacterized protein OS=Carboxylicivirga sp. M1479 OX=2594476 GN=FNN09_06090 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.89HH16 pKa = 3.93GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.12VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.71LSVSSEE47 pKa = 3.93KK48 pKa = 10.19RR49 pKa = 11.84HH50 pKa = 5.73KK51 pKa = 10.69AA52 pKa = 3.17
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.89HH16 pKa = 3.93GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.12VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.71LSVSSEE47 pKa = 3.93KK48 pKa = 10.19RR49 pKa = 11.84HH50 pKa = 5.73KK51 pKa = 10.69AA52 pKa = 3.17
Molecular weight: 6.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1561341 |
34 |
6495 |
381.3 |
42.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.626 ± 0.034 | 0.875 ± 0.013 |
5.818 ± 0.032 | 6.588 ± 0.031 |
4.731 ± 0.026 | 6.632 ± 0.046 |
1.988 ± 0.018 | 7.39 ± 0.035 |
7.009 ± 0.058 | 9.221 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 0.019 | 5.837 ± 0.036 |
3.396 ± 0.021 | 3.736 ± 0.023 |
3.697 ± 0.026 | 6.697 ± 0.035 |
5.305 ± 0.045 | 6.522 ± 0.035 |
1.182 ± 0.014 | 4.242 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
