
Hubei tombus-like virus 30
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.51
Get precalculated fractions of proteins
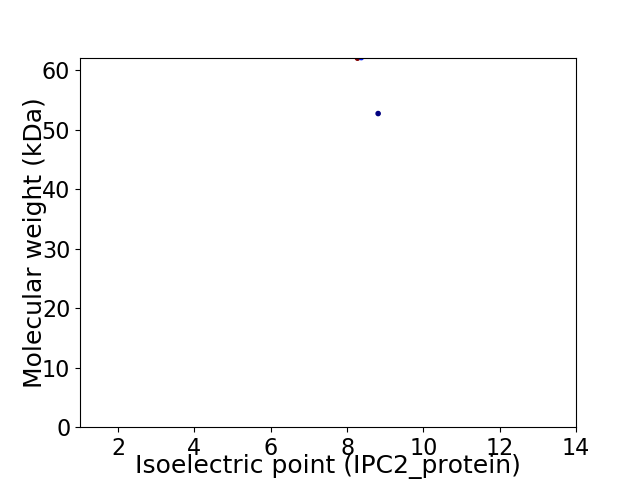
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
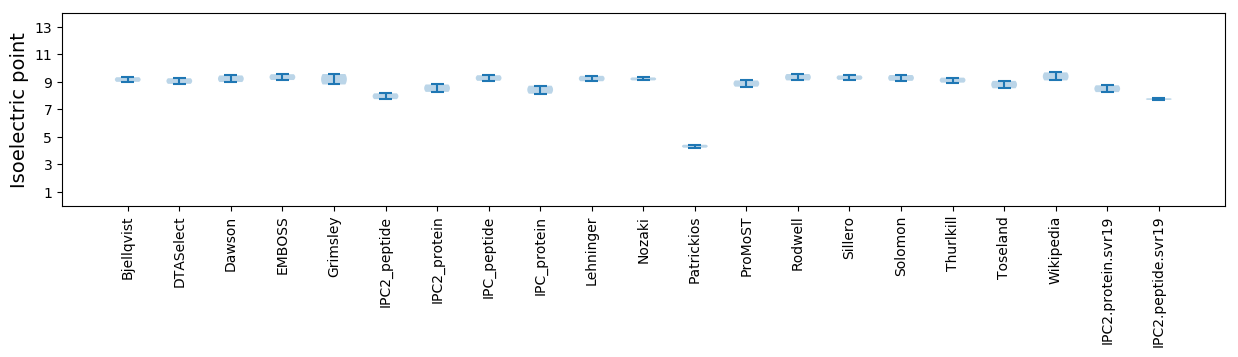
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG52|A0A1L3KG52_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 30 OX=1923278 PE=4 SV=1
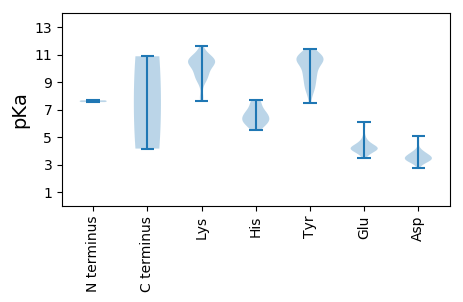
MM1 pKa = 7.54ANSRR5 pKa = 11.84NNDD8 pKa = 3.42PATPGLIHH16 pKa = 7.71KK17 pKa = 7.63MITYY21 pKa = 9.51LFSKK25 pKa = 10.42DD26 pKa = 3.51EE27 pKa = 4.55PEE29 pKa = 4.4TIEE32 pKa = 5.85DD33 pKa = 3.94DD34 pKa = 4.63LEE36 pKa = 5.71SITNAAINLVSLQNSLGEE54 pKa = 4.17IKK56 pKa = 10.64AGATLSEE63 pKa = 4.49VTRR66 pKa = 11.84QEE68 pKa = 4.29QILEE72 pKa = 4.2KK73 pKa = 10.13KK74 pKa = 10.51NKK76 pKa = 9.72IIEE79 pKa = 4.14LLEE82 pKa = 4.25VNDD85 pKa = 4.0QKK87 pKa = 11.58NLEE90 pKa = 4.1PSYY93 pKa = 9.93EE94 pKa = 3.89EE95 pKa = 3.49CRR97 pKa = 11.84AYY99 pKa = 9.84IEE101 pKa = 4.09NRR103 pKa = 11.84VTGKK107 pKa = 10.66LVDD110 pKa = 3.5NTYY113 pKa = 11.01LAFCAALARR122 pKa = 11.84QYY124 pKa = 11.39LSTRR128 pKa = 11.84KK129 pKa = 9.3QVTEE133 pKa = 4.41RR134 pKa = 11.84SANRR138 pKa = 11.84IVQTVLQDD146 pKa = 3.32HH147 pKa = 6.97LEE149 pKa = 4.56GIRR152 pKa = 11.84PKK154 pKa = 10.15WNYY157 pKa = 10.53NIYY160 pKa = 10.76NILKK164 pKa = 9.79HH165 pKa = 6.13NDD167 pKa = 3.39NIGFAEE173 pKa = 3.86NMYY176 pKa = 10.57LKK178 pKa = 10.38FGRR181 pKa = 11.84WVMHH185 pKa = 6.24IPILSWSLPEE195 pKa = 3.85NAQRR199 pKa = 11.84PGITGHH205 pKa = 6.16PFRR208 pKa = 11.84YY209 pKa = 9.51LLSILATLALLYY221 pKa = 10.15ICLSGGSSTAHH232 pKa = 6.11QSTTPAPSITIPNAPPPPLPPLKK255 pKa = 10.57SDD257 pKa = 3.65TPPLNSMSTDD267 pKa = 3.31QNTLSHH273 pKa = 7.33LLTFGSSIANQVSGAVMSTYY293 pKa = 10.72HH294 pKa = 7.62KK295 pKa = 10.16LTTPQKK301 pKa = 9.63EE302 pKa = 4.11NTIKK306 pKa = 10.68EE307 pKa = 4.13LLISLGLVEE316 pKa = 6.07RR317 pKa = 11.84SQPWYY322 pKa = 10.86VRR324 pKa = 11.84LLSWKK329 pKa = 10.25NIAQASTKK337 pKa = 10.62LQDD340 pKa = 3.25SYY342 pKa = 11.25KK343 pKa = 10.39QEE345 pKa = 3.6IHH347 pKa = 6.66PSILSTVDD355 pKa = 2.92TLNQLKK361 pKa = 10.26EE362 pKa = 4.22PLKK365 pKa = 9.68QTYY368 pKa = 8.63TSAKK372 pKa = 9.16EE373 pKa = 4.11HH374 pKa = 5.49TTSAAQKK381 pKa = 10.44LKK383 pKa = 11.14DD384 pKa = 3.75SVANGAGTLSATIPPLMHH402 pKa = 6.7TSRR405 pKa = 11.84SKK407 pKa = 10.26CSSYY411 pKa = 10.11ATSFIEE417 pKa = 4.23NAKK420 pKa = 9.13DD421 pKa = 3.84TIRR424 pKa = 11.84IWNPWRR430 pKa = 11.84AEE432 pKa = 3.84PFTTRR437 pKa = 11.84VRR439 pKa = 11.84RR440 pKa = 11.84ATVSATKK447 pKa = 10.56SLGHH451 pKa = 6.62GCLAMLTLVWEE462 pKa = 4.68TVSSITTYY470 pKa = 10.94LYY472 pKa = 10.35TLLDD476 pKa = 3.6SSVYY480 pKa = 10.75EE481 pKa = 4.0GMPLSMEE488 pKa = 4.27TTASSSQTNPSQPTSTSKK506 pKa = 10.77FFDD509 pKa = 3.46SLIWKK514 pKa = 9.6ALFFLRR520 pKa = 11.84SIIFIKK526 pKa = 10.72LSFAVLGLSTIRR538 pKa = 11.84TRR540 pKa = 11.84TRR542 pKa = 11.84LLCLIPLACVVYY554 pKa = 9.11MVV556 pKa = 4.16
MM1 pKa = 7.54ANSRR5 pKa = 11.84NNDD8 pKa = 3.42PATPGLIHH16 pKa = 7.71KK17 pKa = 7.63MITYY21 pKa = 9.51LFSKK25 pKa = 10.42DD26 pKa = 3.51EE27 pKa = 4.55PEE29 pKa = 4.4TIEE32 pKa = 5.85DD33 pKa = 3.94DD34 pKa = 4.63LEE36 pKa = 5.71SITNAAINLVSLQNSLGEE54 pKa = 4.17IKK56 pKa = 10.64AGATLSEE63 pKa = 4.49VTRR66 pKa = 11.84QEE68 pKa = 4.29QILEE72 pKa = 4.2KK73 pKa = 10.13KK74 pKa = 10.51NKK76 pKa = 9.72IIEE79 pKa = 4.14LLEE82 pKa = 4.25VNDD85 pKa = 4.0QKK87 pKa = 11.58NLEE90 pKa = 4.1PSYY93 pKa = 9.93EE94 pKa = 3.89EE95 pKa = 3.49CRR97 pKa = 11.84AYY99 pKa = 9.84IEE101 pKa = 4.09NRR103 pKa = 11.84VTGKK107 pKa = 10.66LVDD110 pKa = 3.5NTYY113 pKa = 11.01LAFCAALARR122 pKa = 11.84QYY124 pKa = 11.39LSTRR128 pKa = 11.84KK129 pKa = 9.3QVTEE133 pKa = 4.41RR134 pKa = 11.84SANRR138 pKa = 11.84IVQTVLQDD146 pKa = 3.32HH147 pKa = 6.97LEE149 pKa = 4.56GIRR152 pKa = 11.84PKK154 pKa = 10.15WNYY157 pKa = 10.53NIYY160 pKa = 10.76NILKK164 pKa = 9.79HH165 pKa = 6.13NDD167 pKa = 3.39NIGFAEE173 pKa = 3.86NMYY176 pKa = 10.57LKK178 pKa = 10.38FGRR181 pKa = 11.84WVMHH185 pKa = 6.24IPILSWSLPEE195 pKa = 3.85NAQRR199 pKa = 11.84PGITGHH205 pKa = 6.16PFRR208 pKa = 11.84YY209 pKa = 9.51LLSILATLALLYY221 pKa = 10.15ICLSGGSSTAHH232 pKa = 6.11QSTTPAPSITIPNAPPPPLPPLKK255 pKa = 10.57SDD257 pKa = 3.65TPPLNSMSTDD267 pKa = 3.31QNTLSHH273 pKa = 7.33LLTFGSSIANQVSGAVMSTYY293 pKa = 10.72HH294 pKa = 7.62KK295 pKa = 10.16LTTPQKK301 pKa = 9.63EE302 pKa = 4.11NTIKK306 pKa = 10.68EE307 pKa = 4.13LLISLGLVEE316 pKa = 6.07RR317 pKa = 11.84SQPWYY322 pKa = 10.86VRR324 pKa = 11.84LLSWKK329 pKa = 10.25NIAQASTKK337 pKa = 10.62LQDD340 pKa = 3.25SYY342 pKa = 11.25KK343 pKa = 10.39QEE345 pKa = 3.6IHH347 pKa = 6.66PSILSTVDD355 pKa = 2.92TLNQLKK361 pKa = 10.26EE362 pKa = 4.22PLKK365 pKa = 9.68QTYY368 pKa = 8.63TSAKK372 pKa = 9.16EE373 pKa = 4.11HH374 pKa = 5.49TTSAAQKK381 pKa = 10.44LKK383 pKa = 11.14DD384 pKa = 3.75SVANGAGTLSATIPPLMHH402 pKa = 6.7TSRR405 pKa = 11.84SKK407 pKa = 10.26CSSYY411 pKa = 10.11ATSFIEE417 pKa = 4.23NAKK420 pKa = 9.13DD421 pKa = 3.84TIRR424 pKa = 11.84IWNPWRR430 pKa = 11.84AEE432 pKa = 3.84PFTTRR437 pKa = 11.84VRR439 pKa = 11.84RR440 pKa = 11.84ATVSATKK447 pKa = 10.56SLGHH451 pKa = 6.62GCLAMLTLVWEE462 pKa = 4.68TVSSITTYY470 pKa = 10.94LYY472 pKa = 10.35TLLDD476 pKa = 3.6SSVYY480 pKa = 10.75EE481 pKa = 4.0GMPLSMEE488 pKa = 4.27TTASSSQTNPSQPTSTSKK506 pKa = 10.77FFDD509 pKa = 3.46SLIWKK514 pKa = 9.6ALFFLRR520 pKa = 11.84SIIFIKK526 pKa = 10.72LSFAVLGLSTIRR538 pKa = 11.84TRR540 pKa = 11.84TRR542 pKa = 11.84LLCLIPLACVVYY554 pKa = 9.11MVV556 pKa = 4.16
Molecular weight: 61.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG52|A0A1L3KG52_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 30 OX=1923278 PE=4 SV=1
MM1 pKa = 7.67LEE3 pKa = 3.96RR4 pKa = 11.84RR5 pKa = 11.84IQYY8 pKa = 9.94RR9 pKa = 11.84PSIYY13 pKa = 10.52NPGSFYY19 pKa = 11.01YY20 pKa = 10.59YY21 pKa = 8.44PQCSATTVAAIEE33 pKa = 4.2IRR35 pKa = 11.84HH36 pKa = 6.1APVKK40 pKa = 10.16LDD42 pKa = 3.51EE43 pKa = 4.12YY44 pKa = 10.66RR45 pKa = 11.84PEE47 pKa = 4.03HH48 pKa = 6.17FKK50 pKa = 11.3SLTHH54 pKa = 7.06LRR56 pKa = 11.84QFYY59 pKa = 9.78CKK61 pKa = 9.67PGVWSRR67 pKa = 11.84DD68 pKa = 3.35EE69 pKa = 4.61YY70 pKa = 10.78ISQIDD75 pKa = 3.69DD76 pKa = 3.57PAKK79 pKa = 8.97RR80 pKa = 11.84KK81 pKa = 9.78YY82 pKa = 7.48YY83 pKa = 10.51QRR85 pKa = 11.84VADD88 pKa = 3.55QLGFGRR94 pKa = 11.84KK95 pKa = 7.78VSALVCAFTKK105 pKa = 10.56LEE107 pKa = 4.29KK108 pKa = 10.95YY109 pKa = 8.51STSKK113 pKa = 10.95YY114 pKa = 9.36KK115 pKa = 10.66APRR118 pKa = 11.84LIQARR123 pKa = 11.84DD124 pKa = 3.17PSFNLEE130 pKa = 3.63YY131 pKa = 10.84GRR133 pKa = 11.84YY134 pKa = 8.32IKK136 pKa = 10.42PIEE139 pKa = 4.26RR140 pKa = 11.84ALKK143 pKa = 9.09TNVHH147 pKa = 6.5FGKK150 pKa = 9.0GTYY153 pKa = 9.36DD154 pKa = 2.71ICGAKK159 pKa = 9.38IEE161 pKa = 4.18RR162 pKa = 11.84LRR164 pKa = 11.84SKK166 pKa = 9.18WRR168 pKa = 11.84WYY170 pKa = 8.46TEE172 pKa = 4.15CDD174 pKa = 3.12HH175 pKa = 6.51STFDD179 pKa = 3.24AHH181 pKa = 5.85VTVEE185 pKa = 4.0MLKK188 pKa = 10.46LCHH191 pKa = 5.98QFYY194 pKa = 10.64RR195 pKa = 11.84KK196 pKa = 9.55CQGHH200 pKa = 5.53NPYY203 pKa = 9.26MEE205 pKa = 4.24SLARR209 pKa = 11.84RR210 pKa = 11.84TIHH213 pKa = 6.48NKK215 pKa = 7.83GTTRR219 pKa = 11.84YY220 pKa = 8.84GEE222 pKa = 4.21RR223 pKa = 11.84YY224 pKa = 8.91KK225 pKa = 10.46VTGTRR230 pKa = 11.84MSGDD234 pKa = 3.1VDD236 pKa = 3.63TSLGNSLINYY246 pKa = 7.97HH247 pKa = 6.36ILIHH251 pKa = 6.28ALRR254 pKa = 11.84LIGIRR259 pKa = 11.84GDD261 pKa = 3.88AIVNGDD267 pKa = 3.79DD268 pKa = 5.08SIIFTNEE275 pKa = 3.9PIPTDD280 pKa = 3.25KK281 pKa = 10.82YY282 pKa = 10.53VQILRR287 pKa = 11.84QFNMEE292 pKa = 4.4SVVLPSVNNIHH303 pKa = 6.74KK304 pKa = 10.63VEE306 pKa = 4.8FCRR309 pKa = 11.84TRR311 pKa = 11.84LIYY314 pKa = 10.68HH315 pKa = 7.02PDD317 pKa = 3.38KK318 pKa = 11.33NPTLMFDD325 pKa = 3.81PARR328 pKa = 11.84LRR330 pKa = 11.84SIYY333 pKa = 10.59GMTWKK338 pKa = 10.36SYY340 pKa = 9.42PDD342 pKa = 3.5HH343 pKa = 7.64VYY345 pKa = 10.69VKK347 pKa = 9.58YY348 pKa = 11.08LEE350 pKa = 4.69AVSHH354 pKa = 6.81ANSCMNSNSPVGRR367 pKa = 11.84EE368 pKa = 3.71WKK370 pKa = 10.17ALEE373 pKa = 5.06RR374 pKa = 11.84IGKK377 pKa = 8.1QLNLLEE383 pKa = 5.1RR384 pKa = 11.84NIQLEE389 pKa = 4.08LAKK392 pKa = 10.76QNGANRR398 pKa = 11.84PYY400 pKa = 9.74TWDD403 pKa = 3.57YY404 pKa = 10.84LDD406 pKa = 4.12PSITEE411 pKa = 4.6AYY413 pKa = 8.78PNYY416 pKa = 10.61RR417 pKa = 11.84MTDD420 pKa = 3.24KK421 pKa = 10.82QHH423 pKa = 5.75PTVPRR428 pKa = 11.84EE429 pKa = 4.11VTPLDD434 pKa = 3.35HH435 pKa = 7.37LINHH439 pKa = 5.79NTQEE443 pKa = 4.32LLLGPLGYY451 pKa = 10.87
MM1 pKa = 7.67LEE3 pKa = 3.96RR4 pKa = 11.84RR5 pKa = 11.84IQYY8 pKa = 9.94RR9 pKa = 11.84PSIYY13 pKa = 10.52NPGSFYY19 pKa = 11.01YY20 pKa = 10.59YY21 pKa = 8.44PQCSATTVAAIEE33 pKa = 4.2IRR35 pKa = 11.84HH36 pKa = 6.1APVKK40 pKa = 10.16LDD42 pKa = 3.51EE43 pKa = 4.12YY44 pKa = 10.66RR45 pKa = 11.84PEE47 pKa = 4.03HH48 pKa = 6.17FKK50 pKa = 11.3SLTHH54 pKa = 7.06LRR56 pKa = 11.84QFYY59 pKa = 9.78CKK61 pKa = 9.67PGVWSRR67 pKa = 11.84DD68 pKa = 3.35EE69 pKa = 4.61YY70 pKa = 10.78ISQIDD75 pKa = 3.69DD76 pKa = 3.57PAKK79 pKa = 8.97RR80 pKa = 11.84KK81 pKa = 9.78YY82 pKa = 7.48YY83 pKa = 10.51QRR85 pKa = 11.84VADD88 pKa = 3.55QLGFGRR94 pKa = 11.84KK95 pKa = 7.78VSALVCAFTKK105 pKa = 10.56LEE107 pKa = 4.29KK108 pKa = 10.95YY109 pKa = 8.51STSKK113 pKa = 10.95YY114 pKa = 9.36KK115 pKa = 10.66APRR118 pKa = 11.84LIQARR123 pKa = 11.84DD124 pKa = 3.17PSFNLEE130 pKa = 3.63YY131 pKa = 10.84GRR133 pKa = 11.84YY134 pKa = 8.32IKK136 pKa = 10.42PIEE139 pKa = 4.26RR140 pKa = 11.84ALKK143 pKa = 9.09TNVHH147 pKa = 6.5FGKK150 pKa = 9.0GTYY153 pKa = 9.36DD154 pKa = 2.71ICGAKK159 pKa = 9.38IEE161 pKa = 4.18RR162 pKa = 11.84LRR164 pKa = 11.84SKK166 pKa = 9.18WRR168 pKa = 11.84WYY170 pKa = 8.46TEE172 pKa = 4.15CDD174 pKa = 3.12HH175 pKa = 6.51STFDD179 pKa = 3.24AHH181 pKa = 5.85VTVEE185 pKa = 4.0MLKK188 pKa = 10.46LCHH191 pKa = 5.98QFYY194 pKa = 10.64RR195 pKa = 11.84KK196 pKa = 9.55CQGHH200 pKa = 5.53NPYY203 pKa = 9.26MEE205 pKa = 4.24SLARR209 pKa = 11.84RR210 pKa = 11.84TIHH213 pKa = 6.48NKK215 pKa = 7.83GTTRR219 pKa = 11.84YY220 pKa = 8.84GEE222 pKa = 4.21RR223 pKa = 11.84YY224 pKa = 8.91KK225 pKa = 10.46VTGTRR230 pKa = 11.84MSGDD234 pKa = 3.1VDD236 pKa = 3.63TSLGNSLINYY246 pKa = 7.97HH247 pKa = 6.36ILIHH251 pKa = 6.28ALRR254 pKa = 11.84LIGIRR259 pKa = 11.84GDD261 pKa = 3.88AIVNGDD267 pKa = 3.79DD268 pKa = 5.08SIIFTNEE275 pKa = 3.9PIPTDD280 pKa = 3.25KK281 pKa = 10.82YY282 pKa = 10.53VQILRR287 pKa = 11.84QFNMEE292 pKa = 4.4SVVLPSVNNIHH303 pKa = 6.74KK304 pKa = 10.63VEE306 pKa = 4.8FCRR309 pKa = 11.84TRR311 pKa = 11.84LIYY314 pKa = 10.68HH315 pKa = 7.02PDD317 pKa = 3.38KK318 pKa = 11.33NPTLMFDD325 pKa = 3.81PARR328 pKa = 11.84LRR330 pKa = 11.84SIYY333 pKa = 10.59GMTWKK338 pKa = 10.36SYY340 pKa = 9.42PDD342 pKa = 3.5HH343 pKa = 7.64VYY345 pKa = 10.69VKK347 pKa = 9.58YY348 pKa = 11.08LEE350 pKa = 4.69AVSHH354 pKa = 6.81ANSCMNSNSPVGRR367 pKa = 11.84EE368 pKa = 3.71WKK370 pKa = 10.17ALEE373 pKa = 5.06RR374 pKa = 11.84IGKK377 pKa = 8.1QLNLLEE383 pKa = 5.1RR384 pKa = 11.84NIQLEE389 pKa = 4.08LAKK392 pKa = 10.76QNGANRR398 pKa = 11.84PYY400 pKa = 9.74TWDD403 pKa = 3.57YY404 pKa = 10.84LDD406 pKa = 4.12PSITEE411 pKa = 4.6AYY413 pKa = 8.78PNYY416 pKa = 10.61RR417 pKa = 11.84MTDD420 pKa = 3.24KK421 pKa = 10.82QHH423 pKa = 5.75PTVPRR428 pKa = 11.84EE429 pKa = 4.11VTPLDD434 pKa = 3.35HH435 pKa = 7.37LINHH439 pKa = 5.79NTQEE443 pKa = 4.32LLLGPLGYY451 pKa = 10.87
Molecular weight: 52.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007 |
451 |
556 |
503.5 |
57.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.157 ± 0.645 | 1.589 ± 0.248 |
3.774 ± 0.674 | 5.263 ± 0.036 |
2.681 ± 0.123 | 4.27 ± 0.507 |
2.979 ± 0.618 | 6.753 ± 0.197 |
5.76 ± 0.274 | 10.427 ± 1.222 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.986 ± 0.006 | 5.263 ± 0.1 |
5.76 ± 0.003 | 3.774 ± 0.138 |
5.958 ± 1.236 | 8.342 ± 1.573 |
8.044 ± 1.256 | 4.667 ± 0.264 |
1.49 ± 0.097 | 5.065 ± 1.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
