
Warrego virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
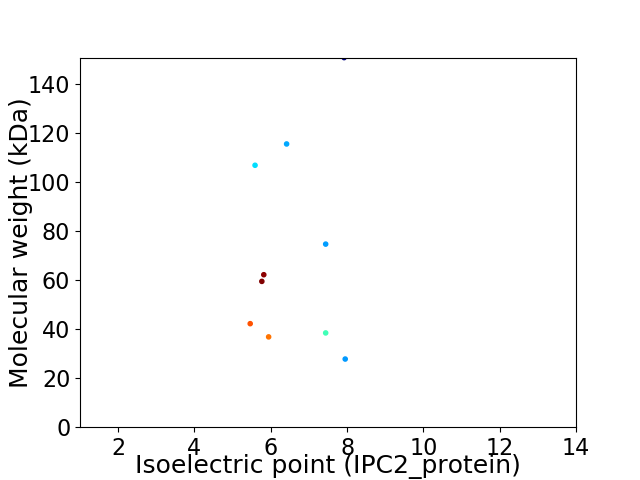
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097I4F1|A0A097I4F1_9REOV Non-structural protein NS3 OS=Warrego virus OX=40062 PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 5.03GKK4 pKa = 8.33KK5 pKa = 9.18TNVPRR10 pKa = 11.84LFTKK14 pKa = 10.3RR15 pKa = 11.84VYY17 pKa = 11.07VLDD20 pKa = 3.76TRR22 pKa = 11.84KK23 pKa = 8.6STICGRR29 pKa = 11.84ISQLAGLPYY38 pKa = 10.54CKK40 pKa = 9.3IEE42 pKa = 4.03SGRR45 pKa = 11.84VVSVTACAQPSDD57 pKa = 3.34RR58 pKa = 11.84GYY60 pKa = 10.83VLEE63 pKa = 4.71IEE65 pKa = 4.69GKK67 pKa = 8.21GAYY70 pKa = 9.72RR71 pKa = 11.84IVDD74 pKa = 3.85GQDD77 pKa = 3.3TISLMIDD84 pKa = 3.35DD85 pKa = 5.33YY86 pKa = 11.13GVSMTTGRR94 pKa = 11.84WEE96 pKa = 3.95EE97 pKa = 4.1YY98 pKa = 9.1AFEE101 pKa = 4.21MVSVRR106 pKa = 11.84PHH108 pKa = 5.22VMKK111 pKa = 10.89VKK113 pKa = 10.14EE114 pKa = 3.96DD115 pKa = 3.26RR116 pKa = 11.84VFKK119 pKa = 10.75NIEE122 pKa = 3.79IKK124 pKa = 10.44HH125 pKa = 5.52GRR127 pKa = 11.84GCAAVPPYY135 pKa = 10.23MRR137 pKa = 11.84VDD139 pKa = 3.78RR140 pKa = 11.84EE141 pKa = 4.22QNSEE145 pKa = 3.76LRR147 pKa = 11.84LPGTEE152 pKa = 3.86RR153 pKa = 11.84SDD155 pKa = 3.4MDD157 pKa = 3.4VSTYY161 pKa = 7.96RR162 pKa = 11.84QKK164 pKa = 10.75RR165 pKa = 11.84RR166 pKa = 11.84EE167 pKa = 3.98EE168 pKa = 4.04RR169 pKa = 11.84EE170 pKa = 3.82GMDD173 pKa = 4.08RR174 pKa = 11.84NTQRR178 pKa = 11.84LQPAHH183 pKa = 6.01MRR185 pKa = 11.84SEE187 pKa = 4.18EE188 pKa = 3.97MDD190 pKa = 3.16RR191 pKa = 11.84SSWSEE196 pKa = 3.51QVNASKK202 pKa = 10.38AQKK205 pKa = 10.29YY206 pKa = 10.17DD207 pKa = 3.25EE208 pKa = 4.82EE209 pKa = 4.46FNKK212 pKa = 10.09FQEE215 pKa = 4.4SVRR218 pKa = 11.84QIEE221 pKa = 5.08LEE223 pKa = 3.96DD224 pKa = 4.39EE225 pKa = 4.55YY226 pKa = 10.9PAGQQDD232 pKa = 5.22DD233 pKa = 5.39DD234 pKa = 4.7NDD236 pKa = 4.48DD237 pKa = 3.73NFGLGLVKK245 pKa = 10.94DD246 pKa = 4.6EE247 pKa = 6.05DD248 pKa = 3.67NTSQKK253 pKa = 10.74KK254 pKa = 10.35GGEE257 pKa = 4.02EE258 pKa = 3.93RR259 pKa = 11.84EE260 pKa = 4.43KK261 pKa = 10.9KK262 pKa = 10.45ASQQDD267 pKa = 3.7DD268 pKa = 3.9FGLDD272 pKa = 3.71FEE274 pKa = 4.88EE275 pKa = 4.9EE276 pKa = 4.32CAKK279 pKa = 10.53FNYY282 pKa = 8.79LTDD285 pKa = 3.77TYY287 pKa = 11.21SEE289 pKa = 4.18LVGKK293 pKa = 10.22IVAKK297 pKa = 10.21NLKK300 pKa = 9.26QLGGKK305 pKa = 9.65SMIFPQLGDD314 pKa = 3.51RR315 pKa = 11.84YY316 pKa = 7.78TEE318 pKa = 3.97KK319 pKa = 10.95LVVLTKK325 pKa = 10.34EE326 pKa = 4.41CNNVPLYY333 pKa = 11.06NIDD336 pKa = 4.26EE337 pKa = 4.22IAKK340 pKa = 8.86EE341 pKa = 3.93YY342 pKa = 10.7RR343 pKa = 11.84FAAIGTCKK351 pKa = 10.19RR352 pKa = 11.84IVMVAKK358 pKa = 10.43GNSFTALPAGAMM370 pKa = 3.88
MM1 pKa = 7.59EE2 pKa = 5.03GKK4 pKa = 8.33KK5 pKa = 9.18TNVPRR10 pKa = 11.84LFTKK14 pKa = 10.3RR15 pKa = 11.84VYY17 pKa = 11.07VLDD20 pKa = 3.76TRR22 pKa = 11.84KK23 pKa = 8.6STICGRR29 pKa = 11.84ISQLAGLPYY38 pKa = 10.54CKK40 pKa = 9.3IEE42 pKa = 4.03SGRR45 pKa = 11.84VVSVTACAQPSDD57 pKa = 3.34RR58 pKa = 11.84GYY60 pKa = 10.83VLEE63 pKa = 4.71IEE65 pKa = 4.69GKK67 pKa = 8.21GAYY70 pKa = 9.72RR71 pKa = 11.84IVDD74 pKa = 3.85GQDD77 pKa = 3.3TISLMIDD84 pKa = 3.35DD85 pKa = 5.33YY86 pKa = 11.13GVSMTTGRR94 pKa = 11.84WEE96 pKa = 3.95EE97 pKa = 4.1YY98 pKa = 9.1AFEE101 pKa = 4.21MVSVRR106 pKa = 11.84PHH108 pKa = 5.22VMKK111 pKa = 10.89VKK113 pKa = 10.14EE114 pKa = 3.96DD115 pKa = 3.26RR116 pKa = 11.84VFKK119 pKa = 10.75NIEE122 pKa = 3.79IKK124 pKa = 10.44HH125 pKa = 5.52GRR127 pKa = 11.84GCAAVPPYY135 pKa = 10.23MRR137 pKa = 11.84VDD139 pKa = 3.78RR140 pKa = 11.84EE141 pKa = 4.22QNSEE145 pKa = 3.76LRR147 pKa = 11.84LPGTEE152 pKa = 3.86RR153 pKa = 11.84SDD155 pKa = 3.4MDD157 pKa = 3.4VSTYY161 pKa = 7.96RR162 pKa = 11.84QKK164 pKa = 10.75RR165 pKa = 11.84RR166 pKa = 11.84EE167 pKa = 3.98EE168 pKa = 4.04RR169 pKa = 11.84EE170 pKa = 3.82GMDD173 pKa = 4.08RR174 pKa = 11.84NTQRR178 pKa = 11.84LQPAHH183 pKa = 6.01MRR185 pKa = 11.84SEE187 pKa = 4.18EE188 pKa = 3.97MDD190 pKa = 3.16RR191 pKa = 11.84SSWSEE196 pKa = 3.51QVNASKK202 pKa = 10.38AQKK205 pKa = 10.29YY206 pKa = 10.17DD207 pKa = 3.25EE208 pKa = 4.82EE209 pKa = 4.46FNKK212 pKa = 10.09FQEE215 pKa = 4.4SVRR218 pKa = 11.84QIEE221 pKa = 5.08LEE223 pKa = 3.96DD224 pKa = 4.39EE225 pKa = 4.55YY226 pKa = 10.9PAGQQDD232 pKa = 5.22DD233 pKa = 5.39DD234 pKa = 4.7NDD236 pKa = 4.48DD237 pKa = 3.73NFGLGLVKK245 pKa = 10.94DD246 pKa = 4.6EE247 pKa = 6.05DD248 pKa = 3.67NTSQKK253 pKa = 10.74KK254 pKa = 10.35GGEE257 pKa = 4.02EE258 pKa = 3.93RR259 pKa = 11.84EE260 pKa = 4.43KK261 pKa = 10.9KK262 pKa = 10.45ASQQDD267 pKa = 3.7DD268 pKa = 3.9FGLDD272 pKa = 3.71FEE274 pKa = 4.88EE275 pKa = 4.9EE276 pKa = 4.32CAKK279 pKa = 10.53FNYY282 pKa = 8.79LTDD285 pKa = 3.77TYY287 pKa = 11.21SEE289 pKa = 4.18LVGKK293 pKa = 10.22IVAKK297 pKa = 10.21NLKK300 pKa = 9.26QLGGKK305 pKa = 9.65SMIFPQLGDD314 pKa = 3.51RR315 pKa = 11.84YY316 pKa = 7.78TEE318 pKa = 3.97KK319 pKa = 10.95LVVLTKK325 pKa = 10.34EE326 pKa = 4.41CNNVPLYY333 pKa = 11.06NIDD336 pKa = 4.26EE337 pKa = 4.22IAKK340 pKa = 8.86EE341 pKa = 3.93YY342 pKa = 10.7RR343 pKa = 11.84FAAIGTCKK351 pKa = 10.19RR352 pKa = 11.84IVMVAKK358 pKa = 10.43GNSFTALPAGAMM370 pKa = 3.88
Molecular weight: 42.22 kDa
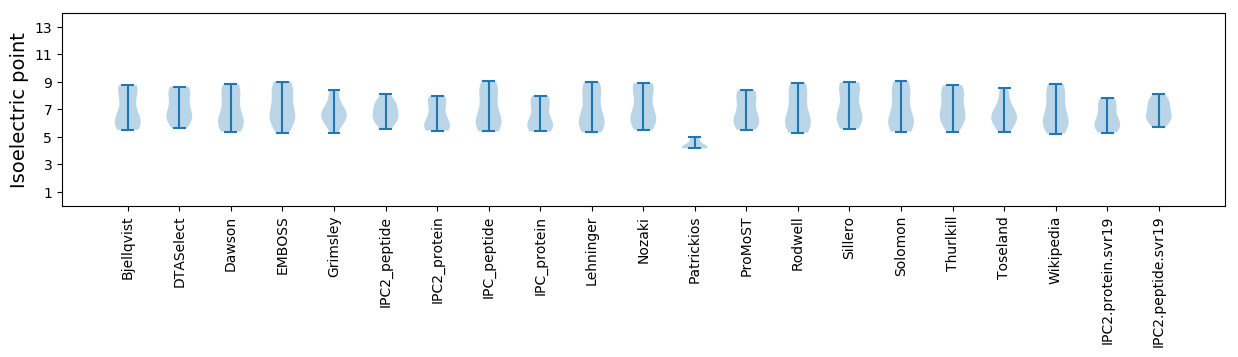
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097I4H0|A0A097I4H0_9REOV Core protein VP4 OS=Warrego virus OX=40062 PE=3 SV=1
MM1 pKa = 7.73LSTLVSRR8 pKa = 11.84CAEE11 pKa = 3.96EE12 pKa = 4.71KK13 pKa = 10.43EE14 pKa = 4.02EE15 pKa = 4.23SRR17 pKa = 11.84RR18 pKa = 11.84LSKK21 pKa = 10.8QFIEE25 pKa = 4.36VDD27 pKa = 3.3LTDD30 pKa = 3.47TMEE33 pKa = 5.6ADD35 pKa = 5.0DD36 pKa = 4.08MNKK39 pKa = 8.71TPSAPPVYY47 pKa = 9.3TPPNAPPNPATVSLNILTNAMSSTTGATNAQKK79 pKa = 10.66EE80 pKa = 4.28EE81 pKa = 3.88KK82 pKa = 10.06AAYY85 pKa = 9.78ASYY88 pKa = 11.15AEE90 pKa = 4.6AMKK93 pKa = 10.83DD94 pKa = 3.55DD95 pKa = 4.1INIKK99 pKa = 9.82IIKK102 pKa = 9.94SRR104 pKa = 11.84VNEE107 pKa = 3.92QIIPRR112 pKa = 11.84LEE114 pKa = 4.83HH115 pKa = 6.52EE116 pKa = 4.41LQSMQRR122 pKa = 11.84RR123 pKa = 11.84KK124 pKa = 10.25KK125 pKa = 9.65ILYY128 pKa = 9.94AVMLISAILALFTSGVTIIKK148 pKa = 9.64DD149 pKa = 3.66LKK151 pKa = 9.44ITLPNSGNATHH162 pKa = 7.77IEE164 pKa = 3.88MPAWVRR170 pKa = 11.84DD171 pKa = 3.45LSVFIGMLNFASMGVALGCAKK192 pKa = 10.21MMDD195 pKa = 3.44GMVRR199 pKa = 11.84SIDD202 pKa = 3.55QVRR205 pKa = 11.84KK206 pKa = 9.84EE207 pKa = 3.85ISKK210 pKa = 10.48KK211 pKa = 9.81KK212 pKa = 10.5AYY214 pKa = 9.62IDD216 pKa = 3.17ATRR219 pKa = 11.84IAFRR223 pKa = 11.84GSVDD227 pKa = 3.83FSKK230 pKa = 11.21LDD232 pKa = 3.36TRR234 pKa = 11.84DD235 pKa = 3.26TEE237 pKa = 4.62RR238 pKa = 11.84YY239 pKa = 9.48AVTPEE244 pKa = 3.69ARR246 pKa = 11.84GLLWW250 pKa = 4.21
MM1 pKa = 7.73LSTLVSRR8 pKa = 11.84CAEE11 pKa = 3.96EE12 pKa = 4.71KK13 pKa = 10.43EE14 pKa = 4.02EE15 pKa = 4.23SRR17 pKa = 11.84RR18 pKa = 11.84LSKK21 pKa = 10.8QFIEE25 pKa = 4.36VDD27 pKa = 3.3LTDD30 pKa = 3.47TMEE33 pKa = 5.6ADD35 pKa = 5.0DD36 pKa = 4.08MNKK39 pKa = 8.71TPSAPPVYY47 pKa = 9.3TPPNAPPNPATVSLNILTNAMSSTTGATNAQKK79 pKa = 10.66EE80 pKa = 4.28EE81 pKa = 3.88KK82 pKa = 10.06AAYY85 pKa = 9.78ASYY88 pKa = 11.15AEE90 pKa = 4.6AMKK93 pKa = 10.83DD94 pKa = 3.55DD95 pKa = 4.1INIKK99 pKa = 9.82IIKK102 pKa = 9.94SRR104 pKa = 11.84VNEE107 pKa = 3.92QIIPRR112 pKa = 11.84LEE114 pKa = 4.83HH115 pKa = 6.52EE116 pKa = 4.41LQSMQRR122 pKa = 11.84RR123 pKa = 11.84KK124 pKa = 10.25KK125 pKa = 9.65ILYY128 pKa = 9.94AVMLISAILALFTSGVTIIKK148 pKa = 9.64DD149 pKa = 3.66LKK151 pKa = 9.44ITLPNSGNATHH162 pKa = 7.77IEE164 pKa = 3.88MPAWVRR170 pKa = 11.84DD171 pKa = 3.45LSVFIGMLNFASMGVALGCAKK192 pKa = 10.21MMDD195 pKa = 3.44GMVRR199 pKa = 11.84SIDD202 pKa = 3.55QVRR205 pKa = 11.84KK206 pKa = 9.84EE207 pKa = 3.85ISKK210 pKa = 10.48KK211 pKa = 9.81KK212 pKa = 10.5AYY214 pKa = 9.62IDD216 pKa = 3.17ATRR219 pKa = 11.84IAFRR223 pKa = 11.84GSVDD227 pKa = 3.83FSKK230 pKa = 11.21LDD232 pKa = 3.36TRR234 pKa = 11.84DD235 pKa = 3.26TEE237 pKa = 4.62RR238 pKa = 11.84YY239 pKa = 9.48AVTPEE244 pKa = 3.69ARR246 pKa = 11.84GLLWW250 pKa = 4.21
Molecular weight: 27.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6212 |
250 |
1308 |
621.2 |
71.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.165 ± 0.62 | 1.062 ± 0.211 |
5.924 ± 0.207 | 7.679 ± 0.558 |
4.089 ± 0.353 | 5.441 ± 0.345 |
2.093 ± 0.236 | 7.308 ± 0.368 |
6.616 ± 0.844 | 8.081 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.912 ± 0.327 | 4.572 ± 0.234 |
3.654 ± 0.297 | 3.719 ± 0.334 |
6.648 ± 0.331 | 5.602 ± 0.316 |
5.425 ± 0.31 | 6.825 ± 0.199 |
1.159 ± 0.17 | 4.024 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
