
Firmicutes bacterium M10-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; unclassified Firmicutes sensu stricto
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
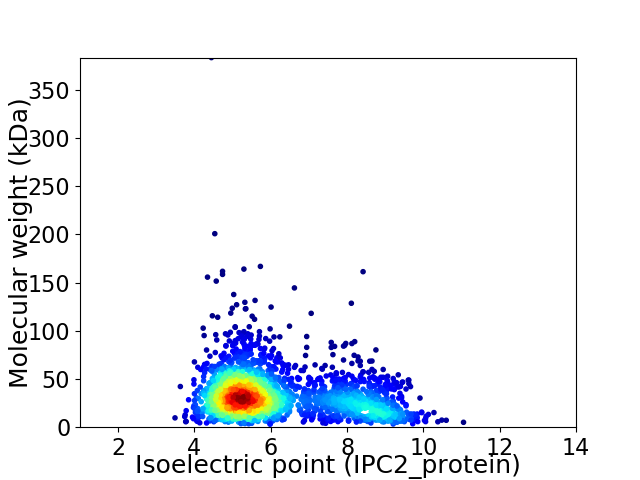
Virtual 2D-PAGE plot for 2256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9LNV1|R9LNV1_9FIRM Orotate phosphoribosyltransferase OS=Firmicutes bacterium M10-2 OX=1235796 GN=pyrE PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 10.57AKK4 pKa = 10.07IALILALTLGLSACSAAPAKK24 pKa = 10.5NQTDD28 pKa = 3.7QPISQQANEE37 pKa = 4.28QFTKK41 pKa = 10.82YY42 pKa = 10.69LNDD45 pKa = 3.56YY46 pKa = 8.12VVEE49 pKa = 4.06MAEE52 pKa = 4.2SDD54 pKa = 4.17FTTMHH59 pKa = 6.19QFFEE63 pKa = 4.42HH64 pKa = 6.96PEE66 pKa = 3.91NYY68 pKa = 10.54GIDD71 pKa = 3.75PEE73 pKa = 4.37KK74 pKa = 11.36VEE76 pKa = 4.15VTLGSIEE83 pKa = 4.06PDD85 pKa = 3.36EE86 pKa = 4.2EE87 pKa = 4.49SKK89 pKa = 11.27ALEE92 pKa = 4.57KK93 pKa = 10.88KK94 pKa = 10.82LSDD97 pKa = 3.52QLDD100 pKa = 3.69SFDD103 pKa = 5.07RR104 pKa = 11.84SSLDD108 pKa = 3.15EE109 pKa = 4.34RR110 pKa = 11.84QQTIYY115 pKa = 11.09DD116 pKa = 3.51QLQYY120 pKa = 11.21QFDD123 pKa = 3.55QSQQEE128 pKa = 4.16EE129 pKa = 4.6EE130 pKa = 4.18EE131 pKa = 4.19KK132 pKa = 10.74FRR134 pKa = 11.84YY135 pKa = 8.59LGNMWSSMNGLPQTLVSYY153 pKa = 10.25FSEE156 pKa = 4.53YY157 pKa = 10.4EE158 pKa = 3.81LRR160 pKa = 11.84QEE162 pKa = 4.42SDD164 pKa = 3.01IAPLIQLINDD174 pKa = 3.6IPRR177 pKa = 11.84YY178 pKa = 7.41TKK180 pKa = 10.38EE181 pKa = 3.51ALTYY185 pKa = 9.92SEE187 pKa = 4.7KK188 pKa = 10.35QAEE191 pKa = 4.66NDD193 pKa = 3.56TLMLDD198 pKa = 3.89YY199 pKa = 10.91EE200 pKa = 4.58DD201 pKa = 5.35TIQSCQEE208 pKa = 3.6ILNSKK213 pKa = 9.34EE214 pKa = 3.72NSAVTAEE221 pKa = 4.02LMKK224 pKa = 10.82DD225 pKa = 3.72VEE227 pKa = 4.57QLDD230 pKa = 4.31LDD232 pKa = 4.03PEE234 pKa = 4.17KK235 pKa = 10.33TIQYY239 pKa = 7.71EE240 pKa = 4.19KK241 pKa = 10.47QIKK244 pKa = 9.65DD245 pKa = 3.43ALDD248 pKa = 3.28TSFFPSYY255 pKa = 11.25QLMIDD260 pKa = 4.17GLTKK264 pKa = 10.76LKK266 pKa = 10.97DD267 pKa = 3.73SIQPVAGMASLEE279 pKa = 4.15NGKK282 pKa = 9.99EE283 pKa = 4.03YY284 pKa = 11.47YY285 pKa = 10.05EE286 pKa = 5.07LLVQQYY292 pKa = 9.05TGTTDD297 pKa = 3.28SIEE300 pKa = 4.34TIKK303 pKa = 11.1KK304 pKa = 9.52NVEE307 pKa = 3.78NEE309 pKa = 3.25IDD311 pKa = 3.97EE312 pKa = 4.93ISTQVSSLLTDD323 pKa = 4.43DD324 pKa = 4.68NYY326 pKa = 11.99DD327 pKa = 3.58MILNYY332 pKa = 9.87MNLKK336 pKa = 9.15TDD338 pKa = 4.13FEE340 pKa = 4.5NVEE343 pKa = 4.71DD344 pKa = 4.21ILVFLNEE351 pKa = 3.77NYY353 pKa = 10.33DD354 pKa = 3.53RR355 pKa = 11.84YY356 pKa = 10.15FPQVDD361 pKa = 3.06AMKK364 pKa = 10.92YY365 pKa = 10.1DD366 pKa = 4.3LQPLADD372 pKa = 4.41DD373 pKa = 3.94QSQEE377 pKa = 4.14GIVAYY382 pKa = 9.38FMIPAVDD389 pKa = 3.3STTDD393 pKa = 2.92YY394 pKa = 10.82RR395 pKa = 11.84IRR397 pKa = 11.84YY398 pKa = 7.39NRR400 pKa = 11.84RR401 pKa = 11.84DD402 pKa = 3.65YY403 pKa = 11.81GDD405 pKa = 4.66DD406 pKa = 3.52PSALSLYY413 pKa = 8.57QTLAHH418 pKa = 6.44EE419 pKa = 5.71GIPGHH424 pKa = 6.54MYY426 pKa = 8.83QAQYY430 pKa = 11.03NKK432 pKa = 9.86EE433 pKa = 3.9HH434 pKa = 5.77FQYY437 pKa = 10.29TIEE440 pKa = 4.18YY441 pKa = 8.72FLSNSGFSEE450 pKa = 5.3GYY452 pKa = 7.93ATYY455 pKa = 11.03VEE457 pKa = 4.12NQSLKK462 pKa = 10.74FLDD465 pKa = 4.9LDD467 pKa = 4.17PDD469 pKa = 4.86LINMYY474 pKa = 10.29VYY476 pKa = 11.06EE477 pKa = 5.1DD478 pKa = 4.21LLNNNYY484 pKa = 9.45ILLMDD489 pKa = 4.0IAVNYY494 pKa = 9.23EE495 pKa = 3.75GLNLEE500 pKa = 4.53EE501 pKa = 4.32FQSTFDD507 pKa = 3.75MFDD510 pKa = 3.21PTALEE515 pKa = 4.27SIYY518 pKa = 10.87DD519 pKa = 3.76QLAGNPGVFMSYY531 pKa = 10.22YY532 pKa = 9.71YY533 pKa = 10.87GYY535 pKa = 11.28NKK537 pKa = 9.84ISNLRR542 pKa = 11.84EE543 pKa = 3.79EE544 pKa = 4.65AKK546 pKa = 10.49EE547 pKa = 3.76ALKK550 pKa = 10.98DD551 pKa = 3.67SFDD554 pKa = 3.89DD555 pKa = 4.23VEE557 pKa = 4.78FNNALLQSGSVNFDD571 pKa = 4.75LIQQNIDD578 pKa = 3.66KK579 pKa = 10.47YY580 pKa = 11.27IKK582 pKa = 10.43DD583 pKa = 3.71NQQ585 pKa = 3.45
MM1 pKa = 7.31KK2 pKa = 10.57AKK4 pKa = 10.07IALILALTLGLSACSAAPAKK24 pKa = 10.5NQTDD28 pKa = 3.7QPISQQANEE37 pKa = 4.28QFTKK41 pKa = 10.82YY42 pKa = 10.69LNDD45 pKa = 3.56YY46 pKa = 8.12VVEE49 pKa = 4.06MAEE52 pKa = 4.2SDD54 pKa = 4.17FTTMHH59 pKa = 6.19QFFEE63 pKa = 4.42HH64 pKa = 6.96PEE66 pKa = 3.91NYY68 pKa = 10.54GIDD71 pKa = 3.75PEE73 pKa = 4.37KK74 pKa = 11.36VEE76 pKa = 4.15VTLGSIEE83 pKa = 4.06PDD85 pKa = 3.36EE86 pKa = 4.2EE87 pKa = 4.49SKK89 pKa = 11.27ALEE92 pKa = 4.57KK93 pKa = 10.88KK94 pKa = 10.82LSDD97 pKa = 3.52QLDD100 pKa = 3.69SFDD103 pKa = 5.07RR104 pKa = 11.84SSLDD108 pKa = 3.15EE109 pKa = 4.34RR110 pKa = 11.84QQTIYY115 pKa = 11.09DD116 pKa = 3.51QLQYY120 pKa = 11.21QFDD123 pKa = 3.55QSQQEE128 pKa = 4.16EE129 pKa = 4.6EE130 pKa = 4.18EE131 pKa = 4.19KK132 pKa = 10.74FRR134 pKa = 11.84YY135 pKa = 8.59LGNMWSSMNGLPQTLVSYY153 pKa = 10.25FSEE156 pKa = 4.53YY157 pKa = 10.4EE158 pKa = 3.81LRR160 pKa = 11.84QEE162 pKa = 4.42SDD164 pKa = 3.01IAPLIQLINDD174 pKa = 3.6IPRR177 pKa = 11.84YY178 pKa = 7.41TKK180 pKa = 10.38EE181 pKa = 3.51ALTYY185 pKa = 9.92SEE187 pKa = 4.7KK188 pKa = 10.35QAEE191 pKa = 4.66NDD193 pKa = 3.56TLMLDD198 pKa = 3.89YY199 pKa = 10.91EE200 pKa = 4.58DD201 pKa = 5.35TIQSCQEE208 pKa = 3.6ILNSKK213 pKa = 9.34EE214 pKa = 3.72NSAVTAEE221 pKa = 4.02LMKK224 pKa = 10.82DD225 pKa = 3.72VEE227 pKa = 4.57QLDD230 pKa = 4.31LDD232 pKa = 4.03PEE234 pKa = 4.17KK235 pKa = 10.33TIQYY239 pKa = 7.71EE240 pKa = 4.19KK241 pKa = 10.47QIKK244 pKa = 9.65DD245 pKa = 3.43ALDD248 pKa = 3.28TSFFPSYY255 pKa = 11.25QLMIDD260 pKa = 4.17GLTKK264 pKa = 10.76LKK266 pKa = 10.97DD267 pKa = 3.73SIQPVAGMASLEE279 pKa = 4.15NGKK282 pKa = 9.99EE283 pKa = 4.03YY284 pKa = 11.47YY285 pKa = 10.05EE286 pKa = 5.07LLVQQYY292 pKa = 9.05TGTTDD297 pKa = 3.28SIEE300 pKa = 4.34TIKK303 pKa = 11.1KK304 pKa = 9.52NVEE307 pKa = 3.78NEE309 pKa = 3.25IDD311 pKa = 3.97EE312 pKa = 4.93ISTQVSSLLTDD323 pKa = 4.43DD324 pKa = 4.68NYY326 pKa = 11.99DD327 pKa = 3.58MILNYY332 pKa = 9.87MNLKK336 pKa = 9.15TDD338 pKa = 4.13FEE340 pKa = 4.5NVEE343 pKa = 4.71DD344 pKa = 4.21ILVFLNEE351 pKa = 3.77NYY353 pKa = 10.33DD354 pKa = 3.53RR355 pKa = 11.84YY356 pKa = 10.15FPQVDD361 pKa = 3.06AMKK364 pKa = 10.92YY365 pKa = 10.1DD366 pKa = 4.3LQPLADD372 pKa = 4.41DD373 pKa = 3.94QSQEE377 pKa = 4.14GIVAYY382 pKa = 9.38FMIPAVDD389 pKa = 3.3STTDD393 pKa = 2.92YY394 pKa = 10.82RR395 pKa = 11.84IRR397 pKa = 11.84YY398 pKa = 7.39NRR400 pKa = 11.84RR401 pKa = 11.84DD402 pKa = 3.65YY403 pKa = 11.81GDD405 pKa = 4.66DD406 pKa = 3.52PSALSLYY413 pKa = 8.57QTLAHH418 pKa = 6.44EE419 pKa = 5.71GIPGHH424 pKa = 6.54MYY426 pKa = 8.83QAQYY430 pKa = 11.03NKK432 pKa = 9.86EE433 pKa = 3.9HH434 pKa = 5.77FQYY437 pKa = 10.29TIEE440 pKa = 4.18YY441 pKa = 8.72FLSNSGFSEE450 pKa = 5.3GYY452 pKa = 7.93ATYY455 pKa = 11.03VEE457 pKa = 4.12NQSLKK462 pKa = 10.74FLDD465 pKa = 4.9LDD467 pKa = 4.17PDD469 pKa = 4.86LINMYY474 pKa = 10.29VYY476 pKa = 11.06EE477 pKa = 5.1DD478 pKa = 4.21LLNNNYY484 pKa = 9.45ILLMDD489 pKa = 4.0IAVNYY494 pKa = 9.23EE495 pKa = 3.75GLNLEE500 pKa = 4.53EE501 pKa = 4.32FQSTFDD507 pKa = 3.75MFDD510 pKa = 3.21PTALEE515 pKa = 4.27SIYY518 pKa = 10.87DD519 pKa = 3.76QLAGNPGVFMSYY531 pKa = 10.22YY532 pKa = 9.71YY533 pKa = 10.87GYY535 pKa = 11.28NKK537 pKa = 9.84ISNLRR542 pKa = 11.84EE543 pKa = 3.79EE544 pKa = 4.65AKK546 pKa = 10.49EE547 pKa = 3.76ALKK550 pKa = 10.98DD551 pKa = 3.67SFDD554 pKa = 3.89DD555 pKa = 4.23VEE557 pKa = 4.78FNNALLQSGSVNFDD571 pKa = 4.75LIQQNIDD578 pKa = 3.66KK579 pKa = 10.47YY580 pKa = 11.27IKK582 pKa = 10.43DD583 pKa = 3.71NQQ585 pKa = 3.45
Molecular weight: 67.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9LWV9|R9LWV9_9FIRM Uncharacterized protein OS=Firmicutes bacterium M10-2 OX=1235796 GN=C815_01577 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.14QPSKK9 pKa = 9.71RR10 pKa = 11.84KK11 pKa = 9.45HH12 pKa = 4.84KK13 pKa = 8.72TVHH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.14QPSKK9 pKa = 9.71RR10 pKa = 11.84KK11 pKa = 9.45HH12 pKa = 4.84KK13 pKa = 8.72TVHH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
690888 |
28 |
3426 |
306.2 |
34.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.643 ± 0.051 | 1.403 ± 0.023 |
5.805 ± 0.044 | 7.362 ± 0.052 |
4.486 ± 0.042 | 6.231 ± 0.044 |
2.125 ± 0.025 | 8.011 ± 0.044 |
7.376 ± 0.043 | 9.298 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.079 ± 0.027 | 4.74 ± 0.037 |
3.37 ± 0.029 | 3.896 ± 0.032 |
3.897 ± 0.044 | 5.769 ± 0.039 |
5.071 ± 0.042 | 6.38 ± 0.045 |
0.982 ± 0.019 | 4.076 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
