
Parabacteroides sp. An277
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; unclassified Parabacteroides
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
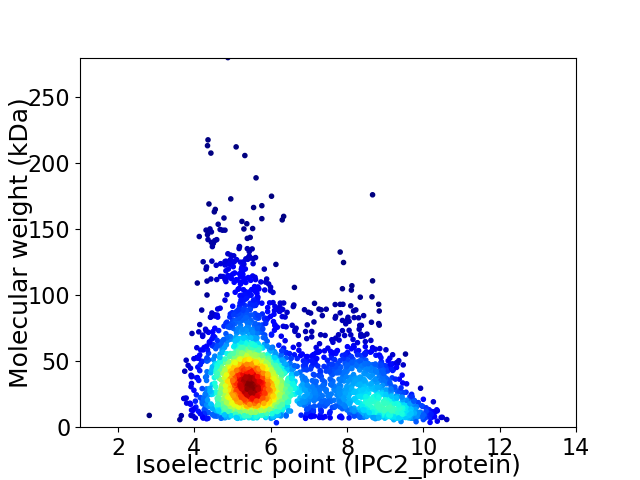
Virtual 2D-PAGE plot for 2949 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4DB68|A0A1Y4DB68_9BACT Uncharacterized protein OS=Parabacteroides sp. An277 OX=1965619 GN=B5F77_02110 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.58KK3 pKa = 10.28NLFYY7 pKa = 10.36TAIFALGMACTFTACSEE24 pKa = 4.03EE25 pKa = 5.13DD26 pKa = 3.75EE27 pKa = 4.97PNPNPSPDD35 pKa = 3.54EE36 pKa = 4.04PTTTSAYY43 pKa = 10.65DD44 pKa = 3.72LDD46 pKa = 4.23YY47 pKa = 10.87TADD50 pKa = 3.8NAAAWGNYY58 pKa = 7.08MYY60 pKa = 9.5QTALLLDD67 pKa = 3.59QDD69 pKa = 3.85ATSLYY74 pKa = 10.23NAWTSDD80 pKa = 3.59YY81 pKa = 10.81EE82 pKa = 4.62GNGPYY87 pKa = 9.29ATIFKK92 pKa = 9.75DD93 pKa = 3.6QTSGAYY99 pKa = 9.58SSPLACIQEE108 pKa = 4.38MVEE111 pKa = 3.83SGMWNIANEE120 pKa = 4.11VGTAKK125 pKa = 10.32IQDD128 pKa = 3.8PYY130 pKa = 8.94NTYY133 pKa = 10.66VSGDD137 pKa = 3.78HH138 pKa = 6.81EE139 pKa = 5.25GGLYY143 pKa = 10.4AVEE146 pKa = 5.26SWFSWHH152 pKa = 7.13SRR154 pKa = 11.84DD155 pKa = 5.48DD156 pKa = 3.47YY157 pKa = 10.93TNNIFSIRR165 pKa = 11.84NTYY168 pKa = 9.12YY169 pKa = 11.15GRR171 pKa = 11.84IDD173 pKa = 4.31DD174 pKa = 4.49NDD176 pKa = 3.24VSKK179 pKa = 10.93IDD181 pKa = 4.23GDD183 pKa = 3.85LSAYY187 pKa = 10.38NSYY190 pKa = 11.57SDD192 pKa = 5.02FDD194 pKa = 4.1DD195 pKa = 5.3AGDD198 pKa = 3.36ISEE201 pKa = 4.74NSLSTFIASINSEE214 pKa = 4.08LDD216 pKa = 3.16EE217 pKa = 4.83TIKK220 pKa = 10.59QQIFAAAKK228 pKa = 9.29AIQAIPQPFRR238 pKa = 11.84NNIDD242 pKa = 3.51SEE244 pKa = 4.5EE245 pKa = 4.15SVAAMDD251 pKa = 4.24ACNTLASTLLNEE263 pKa = 5.01LLPFVNTLSGAEE275 pKa = 3.96YY276 pKa = 10.23TDD278 pKa = 3.59RR279 pKa = 11.84LDD281 pKa = 5.39AIAEE285 pKa = 4.06QFVDD289 pKa = 4.36VVVLPTYY296 pKa = 10.74QDD298 pKa = 3.41LQEE301 pKa = 4.46KK302 pKa = 10.18NRR304 pKa = 11.84TLLDD308 pKa = 3.26VVNQFRR314 pKa = 11.84EE315 pKa = 4.27NPSDD319 pKa = 3.73ANFEE323 pKa = 4.41NVCNAWLEE331 pKa = 4.02ARR333 pKa = 11.84EE334 pKa = 3.96PWEE337 pKa = 3.86KK338 pKa = 11.12SEE340 pKa = 4.51AFLIGPVANWGLDD353 pKa = 3.47PNMDD357 pKa = 3.49SWPLDD362 pKa = 3.3VNAIVQLLEE371 pKa = 4.04SQNWNEE377 pKa = 4.26MQWSGDD383 pKa = 3.61YY384 pKa = 11.36NEE386 pKa = 5.1DD387 pKa = 3.76SEE389 pKa = 5.44TIGAAQNVRR398 pKa = 11.84GYY400 pKa = 8.55HH401 pKa = 4.36TLEE404 pKa = 3.82FLSFKK409 pKa = 10.58NGEE412 pKa = 3.92PRR414 pKa = 11.84KK415 pKa = 10.31VNNN418 pKa = 3.81
MM1 pKa = 7.66KK2 pKa = 10.58KK3 pKa = 10.28NLFYY7 pKa = 10.36TAIFALGMACTFTACSEE24 pKa = 4.03EE25 pKa = 5.13DD26 pKa = 3.75EE27 pKa = 4.97PNPNPSPDD35 pKa = 3.54EE36 pKa = 4.04PTTTSAYY43 pKa = 10.65DD44 pKa = 3.72LDD46 pKa = 4.23YY47 pKa = 10.87TADD50 pKa = 3.8NAAAWGNYY58 pKa = 7.08MYY60 pKa = 9.5QTALLLDD67 pKa = 3.59QDD69 pKa = 3.85ATSLYY74 pKa = 10.23NAWTSDD80 pKa = 3.59YY81 pKa = 10.81EE82 pKa = 4.62GNGPYY87 pKa = 9.29ATIFKK92 pKa = 9.75DD93 pKa = 3.6QTSGAYY99 pKa = 9.58SSPLACIQEE108 pKa = 4.38MVEE111 pKa = 3.83SGMWNIANEE120 pKa = 4.11VGTAKK125 pKa = 10.32IQDD128 pKa = 3.8PYY130 pKa = 8.94NTYY133 pKa = 10.66VSGDD137 pKa = 3.78HH138 pKa = 6.81EE139 pKa = 5.25GGLYY143 pKa = 10.4AVEE146 pKa = 5.26SWFSWHH152 pKa = 7.13SRR154 pKa = 11.84DD155 pKa = 5.48DD156 pKa = 3.47YY157 pKa = 10.93TNNIFSIRR165 pKa = 11.84NTYY168 pKa = 9.12YY169 pKa = 11.15GRR171 pKa = 11.84IDD173 pKa = 4.31DD174 pKa = 4.49NDD176 pKa = 3.24VSKK179 pKa = 10.93IDD181 pKa = 4.23GDD183 pKa = 3.85LSAYY187 pKa = 10.38NSYY190 pKa = 11.57SDD192 pKa = 5.02FDD194 pKa = 4.1DD195 pKa = 5.3AGDD198 pKa = 3.36ISEE201 pKa = 4.74NSLSTFIASINSEE214 pKa = 4.08LDD216 pKa = 3.16EE217 pKa = 4.83TIKK220 pKa = 10.59QQIFAAAKK228 pKa = 9.29AIQAIPQPFRR238 pKa = 11.84NNIDD242 pKa = 3.51SEE244 pKa = 4.5EE245 pKa = 4.15SVAAMDD251 pKa = 4.24ACNTLASTLLNEE263 pKa = 5.01LLPFVNTLSGAEE275 pKa = 3.96YY276 pKa = 10.23TDD278 pKa = 3.59RR279 pKa = 11.84LDD281 pKa = 5.39AIAEE285 pKa = 4.06QFVDD289 pKa = 4.36VVVLPTYY296 pKa = 10.74QDD298 pKa = 3.41LQEE301 pKa = 4.46KK302 pKa = 10.18NRR304 pKa = 11.84TLLDD308 pKa = 3.26VVNQFRR314 pKa = 11.84EE315 pKa = 4.27NPSDD319 pKa = 3.73ANFEE323 pKa = 4.41NVCNAWLEE331 pKa = 4.02ARR333 pKa = 11.84EE334 pKa = 3.96PWEE337 pKa = 3.86KK338 pKa = 11.12SEE340 pKa = 4.51AFLIGPVANWGLDD353 pKa = 3.47PNMDD357 pKa = 3.49SWPLDD362 pKa = 3.3VNAIVQLLEE371 pKa = 4.04SQNWNEE377 pKa = 4.26MQWSGDD383 pKa = 3.61YY384 pKa = 11.36NEE386 pKa = 5.1DD387 pKa = 3.76SEE389 pKa = 5.44TIGAAQNVRR398 pKa = 11.84GYY400 pKa = 8.55HH401 pKa = 4.36TLEE404 pKa = 3.82FLSFKK409 pKa = 10.58NGEE412 pKa = 3.92PRR414 pKa = 11.84KK415 pKa = 10.31VNNN418 pKa = 3.81
Molecular weight: 46.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4CY53|A0A1Y4CY53_9BACT Phenylacetic acid degradation protein OS=Parabacteroides sp. An277 OX=1965619 GN=B5F77_09460 PE=4 SV=1
MM1 pKa = 7.62FEE3 pKa = 4.73LLLIIYY9 pKa = 7.15LVPVIIILSLLWRR22 pKa = 11.84LVLWLVRR29 pKa = 11.84NVLRR33 pKa = 11.84LAGWLLKK40 pKa = 10.2KK41 pKa = 10.62VCVLVWKK48 pKa = 10.52GLLLLVGIFLGRR60 pKa = 11.84CCVNRR65 pKa = 11.84PPDD68 pKa = 3.45SRR70 pKa = 4.23
MM1 pKa = 7.62FEE3 pKa = 4.73LLLIIYY9 pKa = 7.15LVPVIIILSLLWRR22 pKa = 11.84LVLWLVRR29 pKa = 11.84NVLRR33 pKa = 11.84LAGWLLKK40 pKa = 10.2KK41 pKa = 10.62VCVLVWKK48 pKa = 10.52GLLLLVGIFLGRR60 pKa = 11.84CCVNRR65 pKa = 11.84PPDD68 pKa = 3.45SRR70 pKa = 4.23
Molecular weight: 8.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1065573 |
28 |
2472 |
361.3 |
40.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.71 ± 0.044 | 1.21 ± 0.019 |
5.484 ± 0.029 | 6.963 ± 0.043 |
4.436 ± 0.027 | 6.925 ± 0.043 |
2.012 ± 0.024 | 6.412 ± 0.037 |
5.845 ± 0.042 | 9.213 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.649 ± 0.018 | 4.645 ± 0.044 |
3.943 ± 0.027 | 3.749 ± 0.026 |
5.036 ± 0.04 | 5.759 ± 0.032 |
5.675 ± 0.039 | 6.581 ± 0.039 |
1.345 ± 0.02 | 4.41 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
