
Streptomyces indicus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
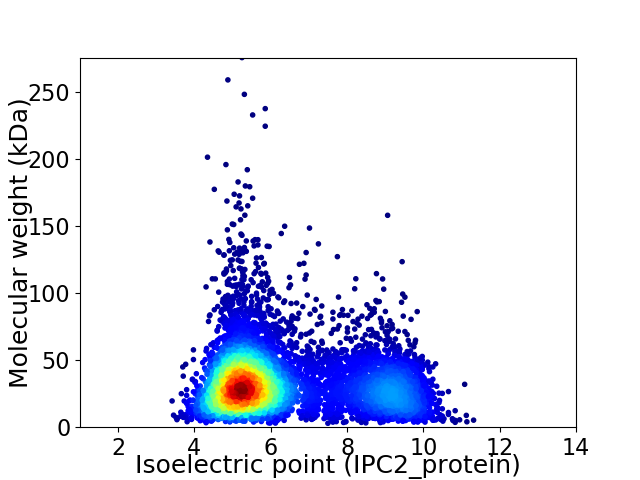
Virtual 2D-PAGE plot for 7387 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
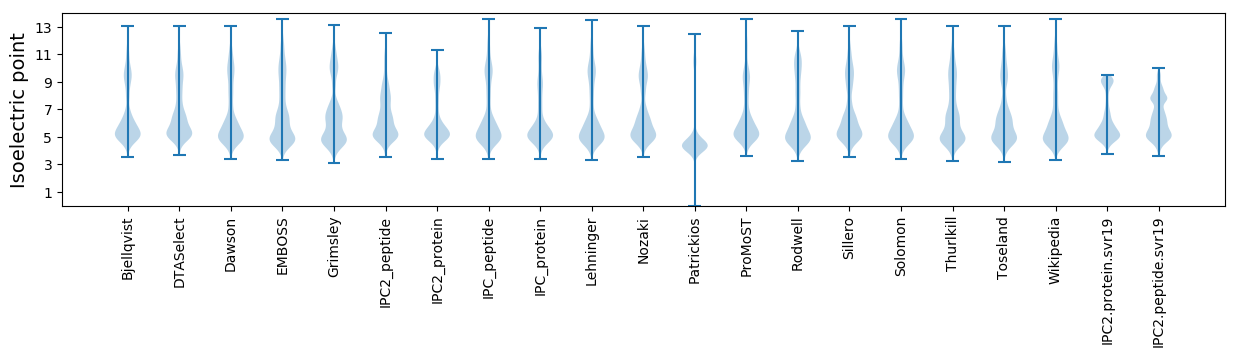
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9DYZ9|A0A1G9DYZ9_9ACTN SsrA-binding protein OS=Streptomyces indicus OX=417292 GN=smpB PE=3 SV=1
MM1 pKa = 7.59SGTDD5 pKa = 3.56GNGVPGYY12 pKa = 10.36GAGDD16 pKa = 4.36GYY18 pKa = 11.48GSGGHH23 pKa = 5.58VSNGQTGEE31 pKa = 3.78GSYY34 pKa = 11.72GDD36 pKa = 4.04GTGGPYY42 pKa = 10.68GDD44 pKa = 4.25GTGGSYY50 pKa = 11.24GDD52 pKa = 4.01GAGAPYY58 pKa = 10.68EE59 pKa = 4.33NGGGAAYY66 pKa = 9.96GHH68 pKa = 6.78GGWQPTSQGDD78 pKa = 3.76YY79 pKa = 11.1DD80 pKa = 4.09GDD82 pKa = 3.52ATAFVALPEE91 pKa = 4.43GLADD95 pKa = 4.04TPLAAPGGNFVPPQISVVPGAAADD119 pKa = 3.78PAATGTFVIPSLVPQMGADD138 pKa = 3.47SAHH141 pKa = 6.41AQPASSPVPSAPAPAPAPVQWPEE164 pKa = 3.88PNAPQQPAPPHH175 pKa = 5.3EE176 pKa = 5.58AGMTGQWSFTEE187 pKa = 4.31AAAQAGPAGQPEE199 pKa = 4.6PVPDD203 pKa = 3.93GQAAQASHH211 pKa = 6.7SPQGNGPHH219 pKa = 7.06ASGPHH224 pKa = 5.85ANVTGQWSIPLAQGDD239 pKa = 4.33LPDD242 pKa = 3.83EE243 pKa = 4.68SGEE246 pKa = 4.29FTSSLADD253 pKa = 3.18QWADD257 pKa = 3.7RR258 pKa = 11.84APATLPGGAPAPWAHH273 pKa = 7.05LVPQDD278 pKa = 4.11EE279 pKa = 4.53EE280 pKa = 4.37QAGQPAQTPHH290 pKa = 6.95GAQEE294 pKa = 3.93PQPAQVGPAEE304 pKa = 4.21AAAAGPEE311 pKa = 4.09PVAAQPEE318 pKa = 4.59DD319 pKa = 3.58PAVPDD324 pKa = 3.85QEE326 pKa = 4.33PQAAEE331 pKa = 4.06APEE334 pKa = 4.06QTDD337 pKa = 3.07QTEE340 pKa = 4.14QTDD343 pKa = 3.54QTEE346 pKa = 4.16QTEE349 pKa = 4.57PARR352 pKa = 11.84QADD355 pKa = 3.71PADD358 pKa = 4.26RR359 pKa = 11.84PEE361 pKa = 4.38QPDD364 pKa = 3.35EE365 pKa = 4.07TAEE368 pKa = 4.07SAEE371 pKa = 4.09AGRR374 pKa = 11.84DD375 pKa = 3.75TEE377 pKa = 4.06EE378 pKa = 3.97SAAFFADD385 pKa = 3.66EE386 pKa = 5.0HH387 pKa = 6.56PLGSYY392 pKa = 7.74VLRR395 pKa = 11.84VNGTDD400 pKa = 4.45RR401 pKa = 11.84PVTDD405 pKa = 2.98AWIGEE410 pKa = 4.15SLLYY414 pKa = 10.18VLRR417 pKa = 11.84EE418 pKa = 3.94RR419 pKa = 11.84LGLAGAKK426 pKa = 9.72DD427 pKa = 3.75GCSQGEE433 pKa = 4.32CGACAVQVDD442 pKa = 3.9GRR444 pKa = 11.84LVASCLVPAATAAGSEE460 pKa = 4.08VRR462 pKa = 11.84TVEE465 pKa = 4.06GLAADD470 pKa = 4.65GQPSDD475 pKa = 3.76VQRR478 pKa = 11.84ALAASCAVQCGFCVPGMAMTVHH500 pKa = 7.27DD501 pKa = 5.21LLEE504 pKa = 4.93GNPQPTEE511 pKa = 3.98RR512 pKa = 11.84EE513 pKa = 3.93TRR515 pKa = 11.84QALCGNLCRR524 pKa = 11.84CSGYY528 pKa = 10.62KK529 pKa = 10.22GVLEE533 pKa = 4.01AVKK536 pKa = 10.63DD537 pKa = 4.19VVAEE541 pKa = 4.12RR542 pKa = 11.84QAAVPDD548 pKa = 3.75EE549 pKa = 5.09DD550 pKa = 4.1DD551 pKa = 3.82PARR554 pKa = 11.84IPHH557 pKa = 5.69QAPPGAGGVNYY568 pKa = 9.37PQDD571 pKa = 3.39GGRR574 pKa = 11.84AA575 pKa = 3.49
MM1 pKa = 7.59SGTDD5 pKa = 3.56GNGVPGYY12 pKa = 10.36GAGDD16 pKa = 4.36GYY18 pKa = 11.48GSGGHH23 pKa = 5.58VSNGQTGEE31 pKa = 3.78GSYY34 pKa = 11.72GDD36 pKa = 4.04GTGGPYY42 pKa = 10.68GDD44 pKa = 4.25GTGGSYY50 pKa = 11.24GDD52 pKa = 4.01GAGAPYY58 pKa = 10.68EE59 pKa = 4.33NGGGAAYY66 pKa = 9.96GHH68 pKa = 6.78GGWQPTSQGDD78 pKa = 3.76YY79 pKa = 11.1DD80 pKa = 4.09GDD82 pKa = 3.52ATAFVALPEE91 pKa = 4.43GLADD95 pKa = 4.04TPLAAPGGNFVPPQISVVPGAAADD119 pKa = 3.78PAATGTFVIPSLVPQMGADD138 pKa = 3.47SAHH141 pKa = 6.41AQPASSPVPSAPAPAPAPVQWPEE164 pKa = 3.88PNAPQQPAPPHH175 pKa = 5.3EE176 pKa = 5.58AGMTGQWSFTEE187 pKa = 4.31AAAQAGPAGQPEE199 pKa = 4.6PVPDD203 pKa = 3.93GQAAQASHH211 pKa = 6.7SPQGNGPHH219 pKa = 7.06ASGPHH224 pKa = 5.85ANVTGQWSIPLAQGDD239 pKa = 4.33LPDD242 pKa = 3.83EE243 pKa = 4.68SGEE246 pKa = 4.29FTSSLADD253 pKa = 3.18QWADD257 pKa = 3.7RR258 pKa = 11.84APATLPGGAPAPWAHH273 pKa = 7.05LVPQDD278 pKa = 4.11EE279 pKa = 4.53EE280 pKa = 4.37QAGQPAQTPHH290 pKa = 6.95GAQEE294 pKa = 3.93PQPAQVGPAEE304 pKa = 4.21AAAAGPEE311 pKa = 4.09PVAAQPEE318 pKa = 4.59DD319 pKa = 3.58PAVPDD324 pKa = 3.85QEE326 pKa = 4.33PQAAEE331 pKa = 4.06APEE334 pKa = 4.06QTDD337 pKa = 3.07QTEE340 pKa = 4.14QTDD343 pKa = 3.54QTEE346 pKa = 4.16QTEE349 pKa = 4.57PARR352 pKa = 11.84QADD355 pKa = 3.71PADD358 pKa = 4.26RR359 pKa = 11.84PEE361 pKa = 4.38QPDD364 pKa = 3.35EE365 pKa = 4.07TAEE368 pKa = 4.07SAEE371 pKa = 4.09AGRR374 pKa = 11.84DD375 pKa = 3.75TEE377 pKa = 4.06EE378 pKa = 3.97SAAFFADD385 pKa = 3.66EE386 pKa = 5.0HH387 pKa = 6.56PLGSYY392 pKa = 7.74VLRR395 pKa = 11.84VNGTDD400 pKa = 4.45RR401 pKa = 11.84PVTDD405 pKa = 2.98AWIGEE410 pKa = 4.15SLLYY414 pKa = 10.18VLRR417 pKa = 11.84EE418 pKa = 3.94RR419 pKa = 11.84LGLAGAKK426 pKa = 9.72DD427 pKa = 3.75GCSQGEE433 pKa = 4.32CGACAVQVDD442 pKa = 3.9GRR444 pKa = 11.84LVASCLVPAATAAGSEE460 pKa = 4.08VRR462 pKa = 11.84TVEE465 pKa = 4.06GLAADD470 pKa = 4.65GQPSDD475 pKa = 3.76VQRR478 pKa = 11.84ALAASCAVQCGFCVPGMAMTVHH500 pKa = 7.27DD501 pKa = 5.21LLEE504 pKa = 4.93GNPQPTEE511 pKa = 3.98RR512 pKa = 11.84EE513 pKa = 3.93TRR515 pKa = 11.84QALCGNLCRR524 pKa = 11.84CSGYY528 pKa = 10.62KK529 pKa = 10.22GVLEE533 pKa = 4.01AVKK536 pKa = 10.63DD537 pKa = 4.19VVAEE541 pKa = 4.12RR542 pKa = 11.84QAAVPDD548 pKa = 3.75EE549 pKa = 5.09DD550 pKa = 4.1DD551 pKa = 3.82PARR554 pKa = 11.84IPHH557 pKa = 5.69QAPPGAGGVNYY568 pKa = 9.37PQDD571 pKa = 3.39GGRR574 pKa = 11.84AA575 pKa = 3.49
Molecular weight: 57.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9FBE4|A0A1G9FBE4_9ACTN Acetoacetate decarboxylase OS=Streptomyces indicus OX=417292 GN=SAMN05421806_11324 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.74GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.74GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2409369 |
28 |
2576 |
326.2 |
34.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.777 ± 0.045 | 0.776 ± 0.009 |
5.817 ± 0.02 | 5.984 ± 0.03 |
2.765 ± 0.016 | 9.551 ± 0.027 |
2.221 ± 0.016 | 3.24 ± 0.02 |
2.512 ± 0.024 | 10.313 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.75 ± 0.013 | 1.744 ± 0.017 |
6.081 ± 0.025 | 2.895 ± 0.016 |
7.748 ± 0.037 | 4.974 ± 0.023 |
5.996 ± 0.03 | 8.236 ± 0.025 |
1.516 ± 0.012 | 2.104 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
