
Plasmodium relictum
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Haemamoeba)
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
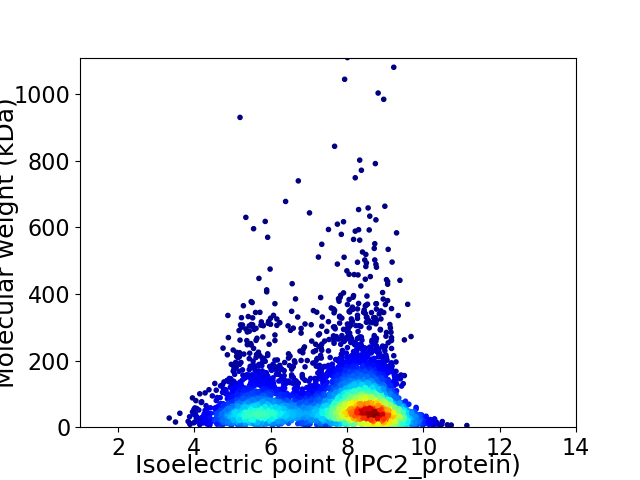
Virtual 2D-PAGE plot for 5136 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J1H0G9|A0A1J1H0G9_PLARL Protein tyrosine phosphatase putative OS=Plasmodium relictum OX=85471 GN=PTP2 PE=4 SV=1
MM1 pKa = 7.66LISVCISQEE10 pKa = 3.94QEE12 pKa = 3.6GNQYY16 pKa = 10.8LFSEE20 pKa = 5.09EE21 pKa = 3.95ISSNVYY27 pKa = 8.27ITEE30 pKa = 4.1NSPNTGNLIDD40 pKa = 4.64TINIEE45 pKa = 3.88KK46 pKa = 10.88DD47 pKa = 3.32EE48 pKa = 4.3ILDD51 pKa = 3.78LEE53 pKa = 4.96NIPSHH58 pKa = 6.97FPIEE62 pKa = 4.37EE63 pKa = 4.23KK64 pKa = 11.04NSDD67 pKa = 4.09SDD69 pKa = 4.24NKK71 pKa = 11.18DD72 pKa = 3.22DD73 pKa = 4.27FFNMSLVDD81 pKa = 4.11TINLEE86 pKa = 3.95KK87 pKa = 10.83SKK89 pKa = 10.95SYY91 pKa = 10.44DD92 pKa = 3.0IKK94 pKa = 11.3NVLDD98 pKa = 4.02EE99 pKa = 4.52VQCSLMDD106 pKa = 3.88SLIDD110 pKa = 3.54PFYY113 pKa = 10.75PSFKK117 pKa = 10.29SEE119 pKa = 4.18KK120 pKa = 10.26LPIHH124 pKa = 6.84IEE126 pKa = 4.18TEE128 pKa = 4.07PSSLMEE134 pKa = 4.37TKK136 pKa = 9.75DD137 pKa = 3.51TEE139 pKa = 4.14HH140 pKa = 7.03KK141 pKa = 10.96YY142 pKa = 10.83SVEE145 pKa = 4.0LEE147 pKa = 4.25NKK149 pKa = 10.19LGEE152 pKa = 4.51TPSTLPDD159 pKa = 3.54SQTQPSNLPLEE170 pKa = 4.72HH171 pKa = 7.1EE172 pKa = 4.1NSIIYY177 pKa = 10.1IEE179 pKa = 4.4TDD181 pKa = 3.31PPSISLMGTKK191 pKa = 8.62YY192 pKa = 9.19TEE194 pKa = 3.46HH195 pKa = 7.15KK196 pKa = 10.74YY197 pKa = 10.5YY198 pKa = 11.11VEE200 pKa = 4.21LQDD203 pKa = 3.79TLGEE207 pKa = 4.29IPSTLPDD214 pKa = 3.37SQTQSFNLLFEE225 pKa = 4.75HH226 pKa = 7.67DD227 pKa = 3.71YY228 pKa = 11.18AINVTHH234 pKa = 6.75GKK236 pKa = 9.69SFSVNNLMNTEE247 pKa = 3.97NVEE250 pKa = 3.98QEE252 pKa = 4.84DD253 pKa = 4.13PVEE256 pKa = 4.07LQNILDD262 pKa = 4.86EE263 pKa = 4.71ILNTLPDD270 pKa = 4.12SLTEE274 pKa = 4.83PIDD277 pKa = 3.62LLSEE281 pKa = 4.45HH282 pKa = 6.84EE283 pKa = 4.37NSTIHH288 pKa = 7.35AEE290 pKa = 4.11AGPSNISLMGTEE302 pKa = 4.21NEE304 pKa = 4.33KK305 pKa = 11.23HH306 pKa = 5.87NDD308 pKa = 3.34SYY310 pKa = 11.68NPQYY314 pKa = 10.62IFNYY318 pKa = 10.48SLVTPNYY325 pKa = 10.28LKK327 pKa = 11.03EE328 pKa = 4.07EE329 pKa = 4.03FLNLLLEE336 pKa = 4.49DD337 pKa = 3.58QCPSINYY344 pKa = 9.63EE345 pKa = 4.52DD346 pKa = 4.05IFPTTSNLMIPEE358 pKa = 4.0NVEE361 pKa = 3.56DD362 pKa = 3.72EE363 pKa = 4.77EE364 pKa = 4.6YY365 pKa = 10.5IKK367 pKa = 10.97LQIIPVDD374 pKa = 4.08FPIEE378 pKa = 4.1KK379 pKa = 9.74PATTINYY386 pKa = 9.16NEE388 pKa = 4.63DD389 pKa = 3.17MSLLDD394 pKa = 4.95EE395 pKa = 4.93IEE397 pKa = 4.25QISS400 pKa = 3.4
MM1 pKa = 7.66LISVCISQEE10 pKa = 3.94QEE12 pKa = 3.6GNQYY16 pKa = 10.8LFSEE20 pKa = 5.09EE21 pKa = 3.95ISSNVYY27 pKa = 8.27ITEE30 pKa = 4.1NSPNTGNLIDD40 pKa = 4.64TINIEE45 pKa = 3.88KK46 pKa = 10.88DD47 pKa = 3.32EE48 pKa = 4.3ILDD51 pKa = 3.78LEE53 pKa = 4.96NIPSHH58 pKa = 6.97FPIEE62 pKa = 4.37EE63 pKa = 4.23KK64 pKa = 11.04NSDD67 pKa = 4.09SDD69 pKa = 4.24NKK71 pKa = 11.18DD72 pKa = 3.22DD73 pKa = 4.27FFNMSLVDD81 pKa = 4.11TINLEE86 pKa = 3.95KK87 pKa = 10.83SKK89 pKa = 10.95SYY91 pKa = 10.44DD92 pKa = 3.0IKK94 pKa = 11.3NVLDD98 pKa = 4.02EE99 pKa = 4.52VQCSLMDD106 pKa = 3.88SLIDD110 pKa = 3.54PFYY113 pKa = 10.75PSFKK117 pKa = 10.29SEE119 pKa = 4.18KK120 pKa = 10.26LPIHH124 pKa = 6.84IEE126 pKa = 4.18TEE128 pKa = 4.07PSSLMEE134 pKa = 4.37TKK136 pKa = 9.75DD137 pKa = 3.51TEE139 pKa = 4.14HH140 pKa = 7.03KK141 pKa = 10.96YY142 pKa = 10.83SVEE145 pKa = 4.0LEE147 pKa = 4.25NKK149 pKa = 10.19LGEE152 pKa = 4.51TPSTLPDD159 pKa = 3.54SQTQPSNLPLEE170 pKa = 4.72HH171 pKa = 7.1EE172 pKa = 4.1NSIIYY177 pKa = 10.1IEE179 pKa = 4.4TDD181 pKa = 3.31PPSISLMGTKK191 pKa = 8.62YY192 pKa = 9.19TEE194 pKa = 3.46HH195 pKa = 7.15KK196 pKa = 10.74YY197 pKa = 10.5YY198 pKa = 11.11VEE200 pKa = 4.21LQDD203 pKa = 3.79TLGEE207 pKa = 4.29IPSTLPDD214 pKa = 3.37SQTQSFNLLFEE225 pKa = 4.75HH226 pKa = 7.67DD227 pKa = 3.71YY228 pKa = 11.18AINVTHH234 pKa = 6.75GKK236 pKa = 9.69SFSVNNLMNTEE247 pKa = 3.97NVEE250 pKa = 3.98QEE252 pKa = 4.84DD253 pKa = 4.13PVEE256 pKa = 4.07LQNILDD262 pKa = 4.86EE263 pKa = 4.71ILNTLPDD270 pKa = 4.12SLTEE274 pKa = 4.83PIDD277 pKa = 3.62LLSEE281 pKa = 4.45HH282 pKa = 6.84EE283 pKa = 4.37NSTIHH288 pKa = 7.35AEE290 pKa = 4.11AGPSNISLMGTEE302 pKa = 4.21NEE304 pKa = 4.33KK305 pKa = 11.23HH306 pKa = 5.87NDD308 pKa = 3.34SYY310 pKa = 11.68NPQYY314 pKa = 10.62IFNYY318 pKa = 10.48SLVTPNYY325 pKa = 10.28LKK327 pKa = 11.03EE328 pKa = 4.07EE329 pKa = 4.03FLNLLLEE336 pKa = 4.49DD337 pKa = 3.58QCPSINYY344 pKa = 9.63EE345 pKa = 4.52DD346 pKa = 4.05IFPTTSNLMIPEE358 pKa = 4.0NVEE361 pKa = 3.56DD362 pKa = 3.72EE363 pKa = 4.77EE364 pKa = 4.6YY365 pKa = 10.5IKK367 pKa = 10.97LQIIPVDD374 pKa = 4.08FPIEE378 pKa = 4.1KK379 pKa = 9.74PATTINYY386 pKa = 9.16NEE388 pKa = 4.63DD389 pKa = 3.17MSLLDD394 pKa = 4.95EE395 pKa = 4.93IEE397 pKa = 4.25QISS400 pKa = 3.4
Molecular weight: 45.69 kDa
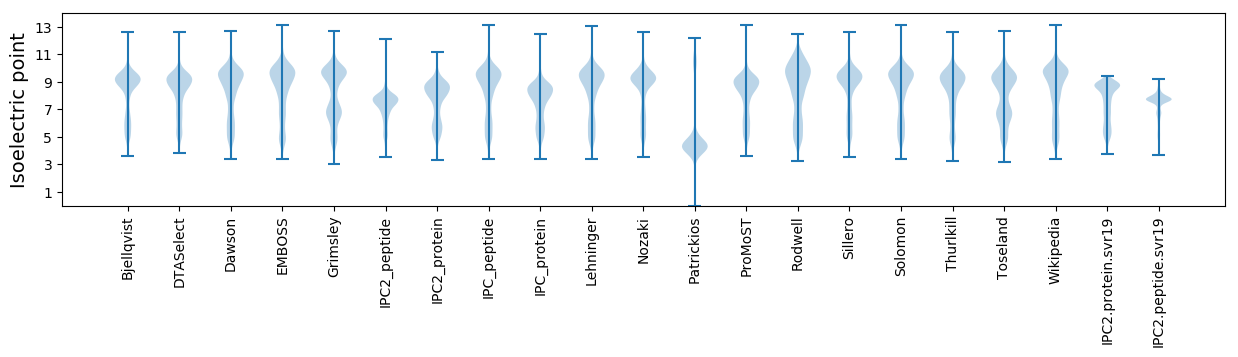
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J1HAA7|A0A1J1HAA7_PLARL Calponin homology domain-containing protein putative OS=Plasmodium relictum OX=85471 GN=PRELSG_1233200 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3624278 |
29 |
9310 |
705.7 |
83.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.851 ± 0.02 | 1.675 ± 0.012 |
5.483 ± 0.024 | 7.944 ± 0.044 |
5.24 ± 0.031 | 2.315 ± 0.022 |
1.728 ± 0.009 | 10.051 ± 0.035 |
13.073 ± 0.055 | 8.569 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.682 ± 0.01 | 13.802 ± 0.102 |
1.659 ± 0.019 | 2.208 ± 0.014 |
2.54 ± 0.016 | 7.133 ± 0.029 |
3.616 ± 0.018 | 3.388 ± 0.019 |
0.451 ± 0.008 | 5.578 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
