
Microbacterium telephonicum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
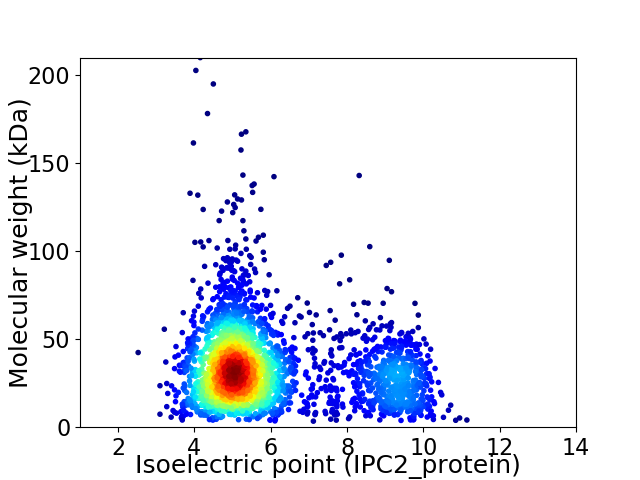
Virtual 2D-PAGE plot for 2897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498BUL3|A0A498BUL3_9MICO Pyruvate formate lyase activating enzyme OS=Microbacterium telephonicum OX=1714841 GN=C7474_2153 PE=3 SV=1
MM1 pKa = 7.65AAQQRR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 4.0VTRR11 pKa = 11.84TLGAGIALIGLIALTACSGGGGEE34 pKa = 4.37VNSDD38 pKa = 3.48YY39 pKa = 11.64GFATAEE45 pKa = 3.94QTADD49 pKa = 3.08SAITVWVDD57 pKa = 3.03AARR60 pKa = 11.84EE61 pKa = 4.07PIAKK65 pKa = 10.02AFEE68 pKa = 4.53ADD70 pKa = 3.69NPDD73 pKa = 2.99VKK75 pKa = 10.97INIEE79 pKa = 4.32TYY81 pKa = 10.34DD82 pKa = 3.71GNAGGSGSFQTKK94 pKa = 9.16VALFDD99 pKa = 3.52QSGEE103 pKa = 3.85GWPDD107 pKa = 3.26VVFSTQTNDD116 pKa = 3.07ASWAAKK122 pKa = 7.14EE123 pKa = 4.38TNGVQAFAAPLDD135 pKa = 3.99KK136 pKa = 11.3GFLDD140 pKa = 4.34QDD142 pKa = 3.78FLDD145 pKa = 4.86GFTSGANDD153 pKa = 3.57PMTVDD158 pKa = 3.56GSVYY162 pKa = 10.81GLRR165 pKa = 11.84NDD167 pKa = 4.41LAPVVLWYY175 pKa = 9.9NQKK178 pKa = 10.66LLDD181 pKa = 3.92EE182 pKa = 5.17FGYY185 pKa = 10.29DD186 pKa = 4.98IPQTWEE192 pKa = 4.09DD193 pKa = 3.79YY194 pKa = 10.6QALGDD199 pKa = 4.06KK200 pKa = 10.29LAAEE204 pKa = 4.41HH205 pKa = 6.84PGYY208 pKa = 10.44ILGSVGDD215 pKa = 4.43SFVGTYY221 pKa = 9.65VYY223 pKa = 11.01YY224 pKa = 10.08WGAQAPIFQLDD235 pKa = 3.57GNTFSSDD242 pKa = 3.12FDD244 pKa = 4.6DD245 pKa = 3.8EE246 pKa = 5.17HH247 pKa = 8.24SVAMTDD253 pKa = 3.56MLDD256 pKa = 3.39HH257 pKa = 6.1MVGNGTLVQDD267 pKa = 4.24SVFSAGFVEE276 pKa = 5.3NYY278 pKa = 9.71ADD280 pKa = 5.27KK281 pKa = 10.47IVAMPGPAWYY291 pKa = 10.33SGALFQNPDD300 pKa = 3.67SLNIPAGEE308 pKa = 4.21IGAADD313 pKa = 4.0PLYY316 pKa = 10.24WDD318 pKa = 4.37GQDD321 pKa = 3.12EE322 pKa = 4.66VTGNVGGGVWYY333 pKa = 10.34ASSHH337 pKa = 5.13SANLEE342 pKa = 3.85AVKK345 pKa = 9.93TFLEE349 pKa = 4.32YY350 pKa = 10.85VISSEE355 pKa = 4.33KK356 pKa = 10.57AVEE359 pKa = 4.03LASGLPAYY367 pKa = 10.37ADD369 pKa = 4.39AASQWLDD376 pKa = 3.41TQASSGYY383 pKa = 10.11YY384 pKa = 10.57DD385 pKa = 4.31GDD387 pKa = 3.4FKK389 pKa = 11.6AAVEE393 pKa = 4.13KK394 pKa = 10.71AAGSVWSGWGFPSFSPEE411 pKa = 3.26TAYY414 pKa = 10.73AAVVVPGLAAGKK426 pKa = 10.07SIADD430 pKa = 3.93VVPDD434 pKa = 3.13WHH436 pKa = 5.75TQILNEE442 pKa = 4.12AQVQGYY448 pKa = 6.84EE449 pKa = 3.98TAEE452 pKa = 3.88
MM1 pKa = 7.65AAQQRR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 4.0VTRR11 pKa = 11.84TLGAGIALIGLIALTACSGGGGEE34 pKa = 4.37VNSDD38 pKa = 3.48YY39 pKa = 11.64GFATAEE45 pKa = 3.94QTADD49 pKa = 3.08SAITVWVDD57 pKa = 3.03AARR60 pKa = 11.84EE61 pKa = 4.07PIAKK65 pKa = 10.02AFEE68 pKa = 4.53ADD70 pKa = 3.69NPDD73 pKa = 2.99VKK75 pKa = 10.97INIEE79 pKa = 4.32TYY81 pKa = 10.34DD82 pKa = 3.71GNAGGSGSFQTKK94 pKa = 9.16VALFDD99 pKa = 3.52QSGEE103 pKa = 3.85GWPDD107 pKa = 3.26VVFSTQTNDD116 pKa = 3.07ASWAAKK122 pKa = 7.14EE123 pKa = 4.38TNGVQAFAAPLDD135 pKa = 3.99KK136 pKa = 11.3GFLDD140 pKa = 4.34QDD142 pKa = 3.78FLDD145 pKa = 4.86GFTSGANDD153 pKa = 3.57PMTVDD158 pKa = 3.56GSVYY162 pKa = 10.81GLRR165 pKa = 11.84NDD167 pKa = 4.41LAPVVLWYY175 pKa = 9.9NQKK178 pKa = 10.66LLDD181 pKa = 3.92EE182 pKa = 5.17FGYY185 pKa = 10.29DD186 pKa = 4.98IPQTWEE192 pKa = 4.09DD193 pKa = 3.79YY194 pKa = 10.6QALGDD199 pKa = 4.06KK200 pKa = 10.29LAAEE204 pKa = 4.41HH205 pKa = 6.84PGYY208 pKa = 10.44ILGSVGDD215 pKa = 4.43SFVGTYY221 pKa = 9.65VYY223 pKa = 11.01YY224 pKa = 10.08WGAQAPIFQLDD235 pKa = 3.57GNTFSSDD242 pKa = 3.12FDD244 pKa = 4.6DD245 pKa = 3.8EE246 pKa = 5.17HH247 pKa = 8.24SVAMTDD253 pKa = 3.56MLDD256 pKa = 3.39HH257 pKa = 6.1MVGNGTLVQDD267 pKa = 4.24SVFSAGFVEE276 pKa = 5.3NYY278 pKa = 9.71ADD280 pKa = 5.27KK281 pKa = 10.47IVAMPGPAWYY291 pKa = 10.33SGALFQNPDD300 pKa = 3.67SLNIPAGEE308 pKa = 4.21IGAADD313 pKa = 4.0PLYY316 pKa = 10.24WDD318 pKa = 4.37GQDD321 pKa = 3.12EE322 pKa = 4.66VTGNVGGGVWYY333 pKa = 10.34ASSHH337 pKa = 5.13SANLEE342 pKa = 3.85AVKK345 pKa = 9.93TFLEE349 pKa = 4.32YY350 pKa = 10.85VISSEE355 pKa = 4.33KK356 pKa = 10.57AVEE359 pKa = 4.03LASGLPAYY367 pKa = 10.37ADD369 pKa = 4.39AASQWLDD376 pKa = 3.41TQASSGYY383 pKa = 10.11YY384 pKa = 10.57DD385 pKa = 4.31GDD387 pKa = 3.4FKK389 pKa = 11.6AAVEE393 pKa = 4.13KK394 pKa = 10.71AAGSVWSGWGFPSFSPEE411 pKa = 3.26TAYY414 pKa = 10.73AAVVVPGLAAGKK426 pKa = 10.07SIADD430 pKa = 3.93VVPDD434 pKa = 3.13WHH436 pKa = 5.75TQILNEE442 pKa = 4.12AQVQGYY448 pKa = 6.84EE449 pKa = 3.98TAEE452 pKa = 3.88
Molecular weight: 47.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498CMH4|A0A498CMH4_9MICO Carbohydrate ABC transporter substrate-binding protein (CUT1 family) OS=Microbacterium telephonicum OX=1714841 GN=C7474_0870 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
950115 |
32 |
2021 |
328.0 |
34.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.359 ± 0.069 | 0.472 ± 0.009 |
6.532 ± 0.043 | 5.305 ± 0.039 |
3.06 ± 0.028 | 9.006 ± 0.039 |
1.972 ± 0.021 | 4.314 ± 0.034 |
1.818 ± 0.034 | 9.978 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.703 ± 0.017 | 1.775 ± 0.027 |
5.494 ± 0.031 | 2.716 ± 0.024 |
7.413 ± 0.055 | 5.151 ± 0.03 |
6.176 ± 0.035 | 9.252 ± 0.047 |
1.545 ± 0.022 | 1.96 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
