
Edhazardia aedis (strain USNM 41457) (Microsporidian parasite)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Microsporidia incertae sedis; Culicosporidae; Edhazardia; Edhazardia aedis
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
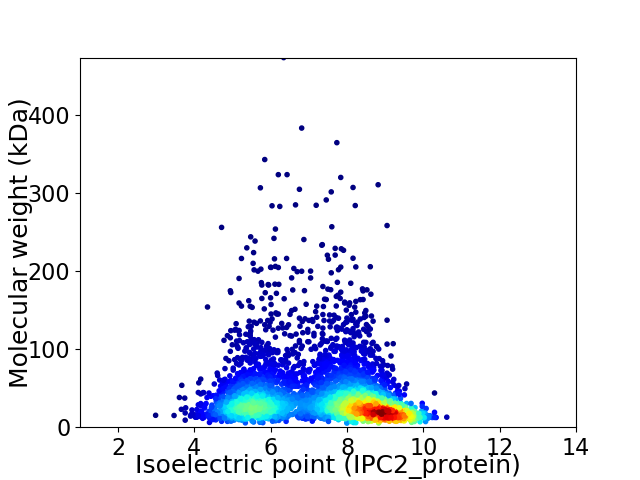
Virtual 2D-PAGE plot for 4189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9DT00|J9DT00_EDHAE RING-type domain-containing protein OS=Edhazardia aedis (strain USNM 41457) OX=1003232 GN=EDEG_01312 PE=4 SV=1
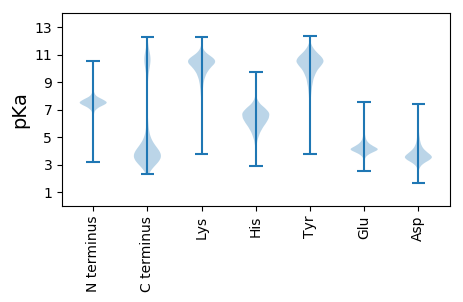
SS1 pKa = 6.83VEE3 pKa = 4.37TPDD6 pKa = 3.58QTGEE10 pKa = 3.92DD11 pKa = 3.69AEE13 pKa = 4.31NPEE16 pKa = 4.24EE17 pKa = 4.23KK18 pKa = 10.53LEE20 pKa = 4.16KK21 pKa = 9.93TSEE24 pKa = 4.19SPEE27 pKa = 4.04NPEE30 pKa = 4.31NNPDD34 pKa = 3.43QAIEE38 pKa = 3.91NAEE41 pKa = 4.05EE42 pKa = 4.0NGKK45 pKa = 9.59SEE47 pKa = 4.38EE48 pKa = 4.22NLDD51 pKa = 3.55QATEE55 pKa = 4.02NTEE58 pKa = 4.18DD59 pKa = 3.94PEE61 pKa = 4.45KK62 pKa = 11.28SEE64 pKa = 5.96DD65 pKa = 3.56NTDD68 pKa = 3.41QTNEE72 pKa = 3.72QSEE75 pKa = 4.42NPEE78 pKa = 4.58DD79 pKa = 4.36NPDD82 pKa = 3.26QAGEE86 pKa = 4.17GNEE89 pKa = 4.14KK90 pKa = 10.52PEE92 pKa = 4.19EE93 pKa = 3.87QCEE96 pKa = 4.54CNDD99 pKa = 3.43HH100 pKa = 6.55PVNEE104 pKa = 4.39LEE106 pKa = 4.27EE107 pKa = 4.61NQDD110 pKa = 3.33QNCQDD115 pKa = 4.34EE116 pKa = 4.83EE117 pKa = 4.81CSCDD121 pKa = 3.31DD122 pKa = 3.79HH123 pKa = 9.25SPNEE127 pKa = 4.33DD128 pKa = 3.92EE129 pKa = 5.31EE130 pKa = 4.98CSCDD134 pKa = 3.37DD135 pKa = 3.79HH136 pKa = 9.25SPNEE140 pKa = 4.33DD141 pKa = 3.92EE142 pKa = 5.31EE143 pKa = 4.98CSCDD147 pKa = 3.37DD148 pKa = 3.79HH149 pKa = 9.38SPNEE153 pKa = 4.49DD154 pKa = 3.8EE155 pKa = 5.53NCEE158 pKa = 4.08CDD160 pKa = 3.89DD161 pKa = 4.01HH162 pKa = 9.73CSDD165 pKa = 3.9EE166 pKa = 5.8DD167 pKa = 4.2EE168 pKa = 4.82NCEE171 pKa = 4.69CDD173 pKa = 3.89DD174 pKa = 4.01HH175 pKa = 9.73CSDD178 pKa = 3.9EE179 pKa = 5.8DD180 pKa = 4.2EE181 pKa = 4.82NCEE184 pKa = 4.95CDD186 pKa = 3.97DD187 pKa = 4.19HH188 pKa = 9.22CPDD191 pKa = 3.63EE192 pKa = 5.93DD193 pKa = 4.54EE194 pKa = 5.21NCEE197 pKa = 4.26CDD199 pKa = 3.89DD200 pKa = 4.01HH201 pKa = 9.73CSDD204 pKa = 3.9EE205 pKa = 5.8DD206 pKa = 4.2EE207 pKa = 4.82NCEE210 pKa = 4.95CDD212 pKa = 3.97DD213 pKa = 4.19HH214 pKa = 9.22CPDD217 pKa = 3.63EE218 pKa = 5.93DD219 pKa = 4.54EE220 pKa = 5.21NCEE223 pKa = 4.09CDD225 pKa = 4.24DD226 pKa = 4.84HH227 pKa = 9.74SGDD230 pKa = 3.79TNEE233 pKa = 4.68TKK235 pKa = 10.55DD236 pKa = 4.18ASGSNNGAEE245 pKa = 4.07NGEE248 pKa = 4.02NLDD251 pKa = 4.01RR252 pKa = 11.84SASHH256 pKa = 6.4PTNPSSNEE264 pKa = 3.76NFQKK268 pKa = 10.69NANNNSPINGSEE280 pKa = 3.88NNPLMKK286 pKa = 10.54NPQDD290 pKa = 3.43QKK292 pKa = 11.73QNLKK296 pKa = 10.17KK297 pKa = 10.05QQNRR301 pKa = 11.84NNSPFCGHH309 pKa = 5.83TPYY312 pKa = 10.64FVFTNRR318 pKa = 11.84CHH320 pKa = 5.5GHH322 pKa = 5.86CGGYY326 pKa = 9.56RR327 pKa = 11.84PKK329 pKa = 10.71NSCKK333 pKa = 10.52
SS1 pKa = 6.83VEE3 pKa = 4.37TPDD6 pKa = 3.58QTGEE10 pKa = 3.92DD11 pKa = 3.69AEE13 pKa = 4.31NPEE16 pKa = 4.24EE17 pKa = 4.23KK18 pKa = 10.53LEE20 pKa = 4.16KK21 pKa = 9.93TSEE24 pKa = 4.19SPEE27 pKa = 4.04NPEE30 pKa = 4.31NNPDD34 pKa = 3.43QAIEE38 pKa = 3.91NAEE41 pKa = 4.05EE42 pKa = 4.0NGKK45 pKa = 9.59SEE47 pKa = 4.38EE48 pKa = 4.22NLDD51 pKa = 3.55QATEE55 pKa = 4.02NTEE58 pKa = 4.18DD59 pKa = 3.94PEE61 pKa = 4.45KK62 pKa = 11.28SEE64 pKa = 5.96DD65 pKa = 3.56NTDD68 pKa = 3.41QTNEE72 pKa = 3.72QSEE75 pKa = 4.42NPEE78 pKa = 4.58DD79 pKa = 4.36NPDD82 pKa = 3.26QAGEE86 pKa = 4.17GNEE89 pKa = 4.14KK90 pKa = 10.52PEE92 pKa = 4.19EE93 pKa = 3.87QCEE96 pKa = 4.54CNDD99 pKa = 3.43HH100 pKa = 6.55PVNEE104 pKa = 4.39LEE106 pKa = 4.27EE107 pKa = 4.61NQDD110 pKa = 3.33QNCQDD115 pKa = 4.34EE116 pKa = 4.83EE117 pKa = 4.81CSCDD121 pKa = 3.31DD122 pKa = 3.79HH123 pKa = 9.25SPNEE127 pKa = 4.33DD128 pKa = 3.92EE129 pKa = 5.31EE130 pKa = 4.98CSCDD134 pKa = 3.37DD135 pKa = 3.79HH136 pKa = 9.25SPNEE140 pKa = 4.33DD141 pKa = 3.92EE142 pKa = 5.31EE143 pKa = 4.98CSCDD147 pKa = 3.37DD148 pKa = 3.79HH149 pKa = 9.38SPNEE153 pKa = 4.49DD154 pKa = 3.8EE155 pKa = 5.53NCEE158 pKa = 4.08CDD160 pKa = 3.89DD161 pKa = 4.01HH162 pKa = 9.73CSDD165 pKa = 3.9EE166 pKa = 5.8DD167 pKa = 4.2EE168 pKa = 4.82NCEE171 pKa = 4.69CDD173 pKa = 3.89DD174 pKa = 4.01HH175 pKa = 9.73CSDD178 pKa = 3.9EE179 pKa = 5.8DD180 pKa = 4.2EE181 pKa = 4.82NCEE184 pKa = 4.95CDD186 pKa = 3.97DD187 pKa = 4.19HH188 pKa = 9.22CPDD191 pKa = 3.63EE192 pKa = 5.93DD193 pKa = 4.54EE194 pKa = 5.21NCEE197 pKa = 4.26CDD199 pKa = 3.89DD200 pKa = 4.01HH201 pKa = 9.73CSDD204 pKa = 3.9EE205 pKa = 5.8DD206 pKa = 4.2EE207 pKa = 4.82NCEE210 pKa = 4.95CDD212 pKa = 3.97DD213 pKa = 4.19HH214 pKa = 9.22CPDD217 pKa = 3.63EE218 pKa = 5.93DD219 pKa = 4.54EE220 pKa = 5.21NCEE223 pKa = 4.09CDD225 pKa = 4.24DD226 pKa = 4.84HH227 pKa = 9.74SGDD230 pKa = 3.79TNEE233 pKa = 4.68TKK235 pKa = 10.55DD236 pKa = 4.18ASGSNNGAEE245 pKa = 4.07NGEE248 pKa = 4.02NLDD251 pKa = 4.01RR252 pKa = 11.84SASHH256 pKa = 6.4PTNPSSNEE264 pKa = 3.76NFQKK268 pKa = 10.69NANNNSPINGSEE280 pKa = 3.88NNPLMKK286 pKa = 10.54NPQDD290 pKa = 3.43QKK292 pKa = 11.73QNLKK296 pKa = 10.17KK297 pKa = 10.05QQNRR301 pKa = 11.84NNSPFCGHH309 pKa = 5.83TPYY312 pKa = 10.64FVFTNRR318 pKa = 11.84CHH320 pKa = 5.5GHH322 pKa = 5.86CGGYY326 pKa = 9.56RR327 pKa = 11.84PKK329 pKa = 10.71NSCKK333 pKa = 10.52
Molecular weight: 37.35 kDa
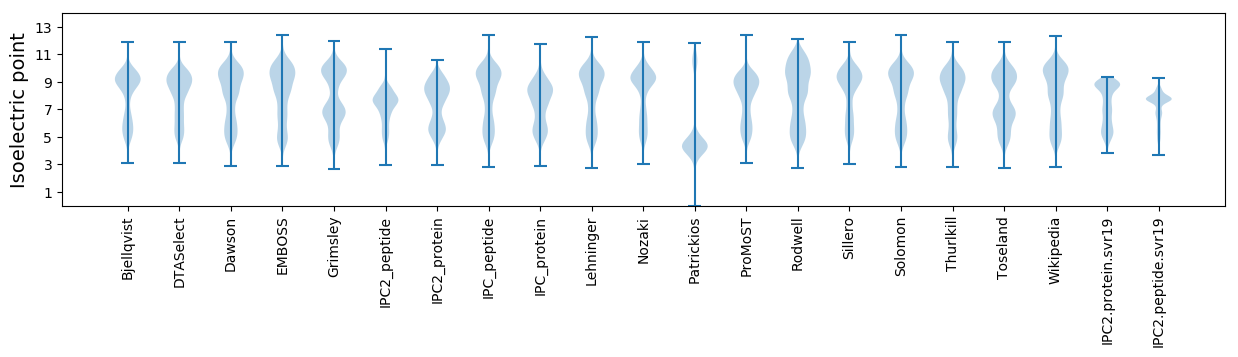
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J8ZW17|J8ZW17_EDHAE Uncharacterized protein OS=Edhazardia aedis (strain USNM 41457) OX=1003232 GN=EDEG_01826 PE=4 SV=1
MM1 pKa = 6.94VFKK4 pKa = 10.68IIARR8 pKa = 11.84EE9 pKa = 4.06SKK11 pKa = 9.63HH12 pKa = 5.13LSRR15 pKa = 11.84RR16 pKa = 11.84EE17 pKa = 3.99SLVQTSHH24 pKa = 6.82KK25 pKa = 10.43FSFLSSILFRR35 pKa = 11.84CLFFKK40 pKa = 10.65NIKK43 pKa = 9.76ILFFNFRR50 pKa = 11.84FFSSLKK56 pKa = 9.29LQIVSSSLPEE66 pKa = 3.82RR67 pKa = 11.84VFVSLDD73 pKa = 3.32LPIIIIITLTNMSRR87 pKa = 11.84NLILRR92 pKa = 11.84PCHH95 pKa = 7.03IYY97 pKa = 9.94EE98 pKa = 4.26QNFF101 pKa = 3.06
MM1 pKa = 6.94VFKK4 pKa = 10.68IIARR8 pKa = 11.84EE9 pKa = 4.06SKK11 pKa = 9.63HH12 pKa = 5.13LSRR15 pKa = 11.84RR16 pKa = 11.84EE17 pKa = 3.99SLVQTSHH24 pKa = 6.82KK25 pKa = 10.43FSFLSSILFRR35 pKa = 11.84CLFFKK40 pKa = 10.65NIKK43 pKa = 9.76ILFFNFRR50 pKa = 11.84FFSSLKK56 pKa = 9.29LQIVSSSLPEE66 pKa = 3.82RR67 pKa = 11.84VFVSLDD73 pKa = 3.32LPIIIIITLTNMSRR87 pKa = 11.84NLILRR92 pKa = 11.84PCHH95 pKa = 7.03IYY97 pKa = 9.94EE98 pKa = 4.26QNFF101 pKa = 3.06
Molecular weight: 12.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1478472 |
45 |
4121 |
352.9 |
41.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.239 ± 0.034 | 1.976 ± 0.017 |
5.783 ± 0.03 | 6.852 ± 0.043 |
5.891 ± 0.041 | 3.001 ± 0.032 |
1.933 ± 0.015 | 9.256 ± 0.036 |
10.545 ± 0.04 | 8.633 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.984 ± 0.015 | 9.999 ± 0.094 |
2.387 ± 0.029 | 3.224 ± 0.022 |
3.293 ± 0.024 | 7.891 ± 0.038 |
4.95 ± 0.025 | 4.51 ± 0.027 |
0.431 ± 0.007 | 4.223 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
