
Torque teno midi virus 10
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
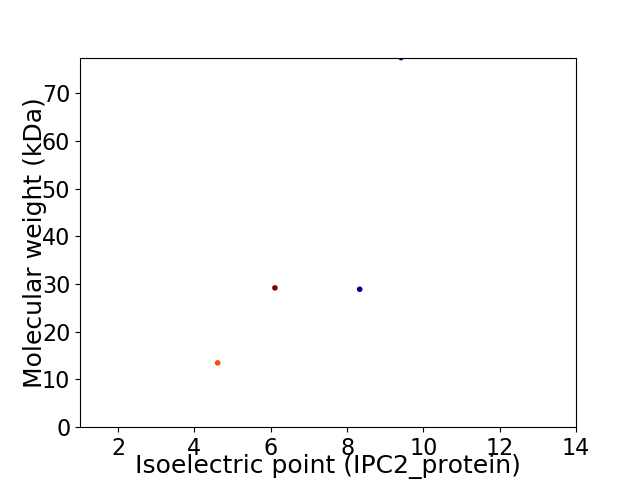
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7VLY8|A7VLY8_9VIRU Capsid protein OS=Torque teno midi virus 10 OX=2065051 PE=3 SV=1
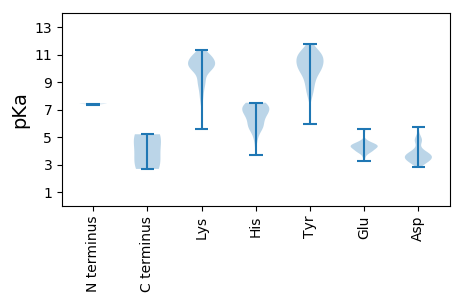
MM1 pKa = 7.43SRR3 pKa = 11.84FYY5 pKa = 10.76TQTKK9 pKa = 10.01YY10 pKa = 11.1NGATLNQMWMSMIADD25 pKa = 3.69SHH27 pKa = 7.49DD28 pKa = 3.11IHH30 pKa = 7.35CDD32 pKa = 3.16CKK34 pKa = 11.08KK35 pKa = 10.52PFAHH39 pKa = 6.99LLDD42 pKa = 4.78NIFPEE47 pKa = 4.38GHH49 pKa = 5.69TDD51 pKa = 3.31RR52 pKa = 11.84NKK54 pKa = 9.94TVAHH58 pKa = 7.21IINRR62 pKa = 11.84DD63 pKa = 3.44YY64 pKa = 11.26KK65 pKa = 8.88EE66 pKa = 4.21CLSGGEE72 pKa = 4.43EE73 pKa = 4.25EE74 pKa = 5.44DD75 pKa = 4.74DD76 pKa = 3.87PGIPVGASAATAAGDD91 pKa = 3.78TEE93 pKa = 4.51IKK95 pKa = 10.71LEE97 pKa = 3.95EE98 pKa = 4.41HH99 pKa = 6.57LEE101 pKa = 3.96EE102 pKa = 4.73DD103 pKa = 3.45AEE105 pKa = 4.38EE106 pKa = 4.24IEE108 pKa = 4.52KK109 pKa = 10.96LLAAAEE115 pKa = 4.12EE116 pKa = 4.46NYY118 pKa = 9.97IRR120 pKa = 5.2
MM1 pKa = 7.43SRR3 pKa = 11.84FYY5 pKa = 10.76TQTKK9 pKa = 10.01YY10 pKa = 11.1NGATLNQMWMSMIADD25 pKa = 3.69SHH27 pKa = 7.49DD28 pKa = 3.11IHH30 pKa = 7.35CDD32 pKa = 3.16CKK34 pKa = 11.08KK35 pKa = 10.52PFAHH39 pKa = 6.99LLDD42 pKa = 4.78NIFPEE47 pKa = 4.38GHH49 pKa = 5.69TDD51 pKa = 3.31RR52 pKa = 11.84NKK54 pKa = 9.94TVAHH58 pKa = 7.21IINRR62 pKa = 11.84DD63 pKa = 3.44YY64 pKa = 11.26KK65 pKa = 8.88EE66 pKa = 4.21CLSGGEE72 pKa = 4.43EE73 pKa = 4.25EE74 pKa = 5.44DD75 pKa = 4.74DD76 pKa = 3.87PGIPVGASAATAAGDD91 pKa = 3.78TEE93 pKa = 4.51IKK95 pKa = 10.71LEE97 pKa = 3.95EE98 pKa = 4.41HH99 pKa = 6.57LEE101 pKa = 3.96EE102 pKa = 4.73DD103 pKa = 3.45AEE105 pKa = 4.38EE106 pKa = 4.24IEE108 pKa = 4.52KK109 pKa = 10.96LLAAAEE115 pKa = 4.12EE116 pKa = 4.46NYY118 pKa = 9.97IRR120 pKa = 5.2
Molecular weight: 13.48 kDa
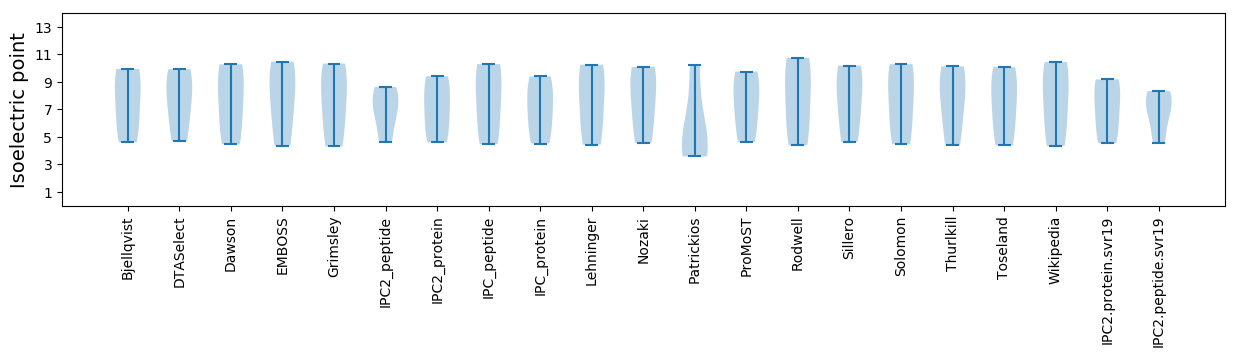
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLY8|A7VLY8_9VIRU Capsid protein OS=Torque teno midi virus 10 OX=2065051 PE=3 SV=1
MM1 pKa = 7.33PFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PWYY13 pKa = 8.27TRR15 pKa = 11.84WGQRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 9.74RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.09RR27 pKa = 11.84NKK29 pKa = 8.86TRR31 pKa = 11.84RR32 pKa = 11.84TFGRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NRR42 pKa = 11.84KK43 pKa = 7.87APRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 7.83LHH53 pKa = 4.89KK54 pKa = 10.07VRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 10.08KK59 pKa = 10.72KK60 pKa = 9.68FLKK63 pKa = 10.15LVQWQPDD70 pKa = 4.08RR71 pKa = 11.84IRR73 pKa = 11.84KK74 pKa = 7.59CTIKK78 pKa = 10.81GRR80 pKa = 11.84GTLVMGAHH88 pKa = 6.8GRR90 pKa = 11.84QMICFTNVKK99 pKa = 10.31DD100 pKa = 3.54RR101 pKa = 11.84LTPSRR106 pKa = 11.84APAGGGFGVEE116 pKa = 4.65RR117 pKa = 11.84ITLAHH122 pKa = 7.34LYY124 pKa = 9.98EE125 pKa = 4.18EE126 pKa = 4.33WKK128 pKa = 10.74ARR130 pKa = 11.84NNVWTATNEE139 pKa = 3.87NTDD142 pKa = 3.24LGRR145 pKa = 11.84YY146 pKa = 5.95TGCKK150 pKa = 8.51ISFYY154 pKa = 10.79RR155 pKa = 11.84HH156 pKa = 6.73LDD158 pKa = 2.84TDD160 pKa = 4.18FIIKK164 pKa = 10.13YY165 pKa = 9.96SRR167 pKa = 11.84NPPFNLEE174 pKa = 4.22KK175 pKa = 10.5YY176 pKa = 10.2SYY178 pKa = 10.41MYY180 pKa = 7.97MQPQEE185 pKa = 4.32LLLGKK190 pKa = 9.62HH191 pKa = 5.6KK192 pKa = 10.73RR193 pKa = 11.84ILLSKK198 pKa = 9.01KK199 pKa = 8.17TNPKK203 pKa = 10.32GKK205 pKa = 10.4LKK207 pKa = 8.36TTLKK211 pKa = 10.34IGPPKK216 pKa = 10.73LLTNKK221 pKa = 9.08WLLQSEE227 pKa = 4.56LAKK230 pKa = 10.26QDD232 pKa = 3.6LVEE235 pKa = 4.53ISAAAASFTYY245 pKa = 8.25PTIGCCNEE253 pKa = 3.28NRR255 pKa = 11.84ILNLYY260 pKa = 9.89SINTEE265 pKa = 4.27FYY267 pKa = 11.07KK268 pKa = 10.61DD269 pKa = 3.64AHH271 pKa = 6.15WMQAHH276 pKa = 7.31AGTNYY281 pKa = 9.14YY282 pKa = 10.17KK283 pKa = 10.23PYY285 pKa = 9.56STIPSDD291 pKa = 4.58LKK293 pKa = 10.7FHH295 pKa = 6.81YY296 pKa = 8.18KK297 pKa = 7.68TQSGGEE303 pKa = 4.14SAINGPDD310 pKa = 3.5SKK312 pKa = 11.17EE313 pKa = 3.74KK314 pKa = 10.9ALTLEE319 pKa = 4.82GGWFQPKK326 pKa = 9.74ILGAYY331 pKa = 9.25KK332 pKa = 8.79ITKK335 pKa = 9.93GSTITQYY342 pKa = 11.76GEE344 pKa = 3.96LPMVLLRR351 pKa = 11.84YY352 pKa = 9.87NPAADD357 pKa = 3.49QGPGNIVTIIDD368 pKa = 4.06LVSGQWQGPYY378 pKa = 9.9PDD380 pKa = 5.72DD381 pKa = 3.63YY382 pKa = 10.83TIKK385 pKa = 10.68DD386 pKa = 3.61VPLWLALNGYY396 pKa = 8.72PNYY399 pKa = 10.43VMQKK403 pKa = 10.0SGDD406 pKa = 3.56KK407 pKa = 11.05GILKK411 pKa = 10.33HH412 pKa = 7.01KK413 pKa = 9.0LICVKK418 pKa = 10.45SAALYY423 pKa = 9.64RR424 pKa = 11.84LPTVSTQKK432 pKa = 9.72IFPIIGVKK440 pKa = 9.76FVNGKK445 pKa = 8.93PPYY448 pKa = 9.0DD449 pKa = 3.56QPLTSDD455 pKa = 3.9MSKK458 pKa = 10.13AWYY461 pKa = 8.61PIYY464 pKa = 10.22EE465 pKa = 4.31HH466 pKa = 5.75QQEE469 pKa = 4.31PLNDD473 pKa = 3.66IVCCGPLVPKK483 pKa = 10.24FYY485 pKa = 10.96NQTNSTWEE493 pKa = 3.56LQYY496 pKa = 10.64FYY498 pKa = 11.03RR499 pKa = 11.84FYY501 pKa = 10.8FKK503 pKa = 10.24WGGPEE508 pKa = 3.74IGDD511 pKa = 3.82PTVDD515 pKa = 3.66DD516 pKa = 5.13PKK518 pKa = 10.97TKK520 pKa = 10.47GQATTDD526 pKa = 3.41YY527 pKa = 10.04TMPRR531 pKa = 11.84SIQVFNPLKK540 pKa = 10.77NKK542 pKa = 9.32TEE544 pKa = 4.66SILHH548 pKa = 5.88PWDD551 pKa = 3.57FRR553 pKa = 11.84RR554 pKa = 11.84GIVTKK559 pKa = 10.42RR560 pKa = 11.84ALEE563 pKa = 3.96RR564 pKa = 11.84MCQNLEE570 pKa = 4.0TDD572 pKa = 3.7SNFYY576 pKa = 10.38PDD578 pKa = 4.76GEE580 pKa = 4.53TPKK583 pKa = 10.4KK584 pKa = 10.08KK585 pKa = 10.45KK586 pKa = 9.18KK587 pKa = 5.61TTCEE591 pKa = 3.44LRR593 pKa = 11.84NPQEE597 pKa = 4.24EE598 pKa = 4.21IKK600 pKa = 10.47EE601 pKa = 4.15VQSCILSLCQTEE613 pKa = 4.37TSEE616 pKa = 5.61EE617 pKa = 4.03EE618 pKa = 4.37DD619 pKa = 3.94PNNLQQLIHH628 pKa = 5.91KK629 pKa = 9.86QKK631 pKa = 10.85LKK633 pKa = 10.14QQQLKK638 pKa = 10.82HH639 pKa = 6.35NILVLLTDD647 pKa = 5.09LKK649 pKa = 10.33TKK651 pKa = 10.54QNSLRR656 pKa = 11.84LQTGNLQQ663 pKa = 3.29
MM1 pKa = 7.33PFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PWYY13 pKa = 8.27TRR15 pKa = 11.84WGQRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 9.74RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.09RR27 pKa = 11.84NKK29 pKa = 8.86TRR31 pKa = 11.84RR32 pKa = 11.84TFGRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NRR42 pKa = 11.84KK43 pKa = 7.87APRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 7.83LHH53 pKa = 4.89KK54 pKa = 10.07VRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 10.08KK59 pKa = 10.72KK60 pKa = 9.68FLKK63 pKa = 10.15LVQWQPDD70 pKa = 4.08RR71 pKa = 11.84IRR73 pKa = 11.84KK74 pKa = 7.59CTIKK78 pKa = 10.81GRR80 pKa = 11.84GTLVMGAHH88 pKa = 6.8GRR90 pKa = 11.84QMICFTNVKK99 pKa = 10.31DD100 pKa = 3.54RR101 pKa = 11.84LTPSRR106 pKa = 11.84APAGGGFGVEE116 pKa = 4.65RR117 pKa = 11.84ITLAHH122 pKa = 7.34LYY124 pKa = 9.98EE125 pKa = 4.18EE126 pKa = 4.33WKK128 pKa = 10.74ARR130 pKa = 11.84NNVWTATNEE139 pKa = 3.87NTDD142 pKa = 3.24LGRR145 pKa = 11.84YY146 pKa = 5.95TGCKK150 pKa = 8.51ISFYY154 pKa = 10.79RR155 pKa = 11.84HH156 pKa = 6.73LDD158 pKa = 2.84TDD160 pKa = 4.18FIIKK164 pKa = 10.13YY165 pKa = 9.96SRR167 pKa = 11.84NPPFNLEE174 pKa = 4.22KK175 pKa = 10.5YY176 pKa = 10.2SYY178 pKa = 10.41MYY180 pKa = 7.97MQPQEE185 pKa = 4.32LLLGKK190 pKa = 9.62HH191 pKa = 5.6KK192 pKa = 10.73RR193 pKa = 11.84ILLSKK198 pKa = 9.01KK199 pKa = 8.17TNPKK203 pKa = 10.32GKK205 pKa = 10.4LKK207 pKa = 8.36TTLKK211 pKa = 10.34IGPPKK216 pKa = 10.73LLTNKK221 pKa = 9.08WLLQSEE227 pKa = 4.56LAKK230 pKa = 10.26QDD232 pKa = 3.6LVEE235 pKa = 4.53ISAAAASFTYY245 pKa = 8.25PTIGCCNEE253 pKa = 3.28NRR255 pKa = 11.84ILNLYY260 pKa = 9.89SINTEE265 pKa = 4.27FYY267 pKa = 11.07KK268 pKa = 10.61DD269 pKa = 3.64AHH271 pKa = 6.15WMQAHH276 pKa = 7.31AGTNYY281 pKa = 9.14YY282 pKa = 10.17KK283 pKa = 10.23PYY285 pKa = 9.56STIPSDD291 pKa = 4.58LKK293 pKa = 10.7FHH295 pKa = 6.81YY296 pKa = 8.18KK297 pKa = 7.68TQSGGEE303 pKa = 4.14SAINGPDD310 pKa = 3.5SKK312 pKa = 11.17EE313 pKa = 3.74KK314 pKa = 10.9ALTLEE319 pKa = 4.82GGWFQPKK326 pKa = 9.74ILGAYY331 pKa = 9.25KK332 pKa = 8.79ITKK335 pKa = 9.93GSTITQYY342 pKa = 11.76GEE344 pKa = 3.96LPMVLLRR351 pKa = 11.84YY352 pKa = 9.87NPAADD357 pKa = 3.49QGPGNIVTIIDD368 pKa = 4.06LVSGQWQGPYY378 pKa = 9.9PDD380 pKa = 5.72DD381 pKa = 3.63YY382 pKa = 10.83TIKK385 pKa = 10.68DD386 pKa = 3.61VPLWLALNGYY396 pKa = 8.72PNYY399 pKa = 10.43VMQKK403 pKa = 10.0SGDD406 pKa = 3.56KK407 pKa = 11.05GILKK411 pKa = 10.33HH412 pKa = 7.01KK413 pKa = 9.0LICVKK418 pKa = 10.45SAALYY423 pKa = 9.64RR424 pKa = 11.84LPTVSTQKK432 pKa = 9.72IFPIIGVKK440 pKa = 9.76FVNGKK445 pKa = 8.93PPYY448 pKa = 9.0DD449 pKa = 3.56QPLTSDD455 pKa = 3.9MSKK458 pKa = 10.13AWYY461 pKa = 8.61PIYY464 pKa = 10.22EE465 pKa = 4.31HH466 pKa = 5.75QQEE469 pKa = 4.31PLNDD473 pKa = 3.66IVCCGPLVPKK483 pKa = 10.24FYY485 pKa = 10.96NQTNSTWEE493 pKa = 3.56LQYY496 pKa = 10.64FYY498 pKa = 11.03RR499 pKa = 11.84FYY501 pKa = 10.8FKK503 pKa = 10.24WGGPEE508 pKa = 3.74IGDD511 pKa = 3.82PTVDD515 pKa = 3.66DD516 pKa = 5.13PKK518 pKa = 10.97TKK520 pKa = 10.47GQATTDD526 pKa = 3.41YY527 pKa = 10.04TMPRR531 pKa = 11.84SIQVFNPLKK540 pKa = 10.77NKK542 pKa = 9.32TEE544 pKa = 4.66SILHH548 pKa = 5.88PWDD551 pKa = 3.57FRR553 pKa = 11.84RR554 pKa = 11.84GIVTKK559 pKa = 10.42RR560 pKa = 11.84ALEE563 pKa = 3.96RR564 pKa = 11.84MCQNLEE570 pKa = 4.0TDD572 pKa = 3.7SNFYY576 pKa = 10.38PDD578 pKa = 4.76GEE580 pKa = 4.53TPKK583 pKa = 10.4KK584 pKa = 10.08KK585 pKa = 10.45KK586 pKa = 9.18KK587 pKa = 5.61TTCEE591 pKa = 3.44LRR593 pKa = 11.84NPQEE597 pKa = 4.24EE598 pKa = 4.21IKK600 pKa = 10.47EE601 pKa = 4.15VQSCILSLCQTEE613 pKa = 4.37TSEE616 pKa = 5.61EE617 pKa = 4.03EE618 pKa = 4.37DD619 pKa = 3.94PNNLQQLIHH628 pKa = 5.91KK629 pKa = 9.86QKK631 pKa = 10.85LKK633 pKa = 10.14QQQLKK638 pKa = 10.82HH639 pKa = 6.35NILVLLTDD647 pKa = 5.09LKK649 pKa = 10.33TKK651 pKa = 10.54QNSLRR656 pKa = 11.84LQTGNLQQ663 pKa = 3.29
Molecular weight: 77.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1297 |
120 |
663 |
324.3 |
37.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.708 ± 1.867 | 1.928 ± 0.095 |
4.703 ± 0.587 | 6.862 ± 1.541 |
2.93 ± 0.376 | 5.32 ± 0.715 |
3.007 ± 0.677 | 5.628 ± 0.258 |
9.098 ± 0.957 | 8.096 ± 0.819 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.251 | 5.012 ± 0.614 |
6.322 ± 0.736 | 5.166 ± 0.49 |
6.322 ± 0.915 | 5.628 ± 1.619 |
7.17 ± 0.459 | 2.39 ± 0.709 |
1.619 ± 0.52 | 4.086 ± 0.688 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
