
Phaeocystidibacter marisrubri
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Schleiferiaceae; Phaeocystidibacter
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
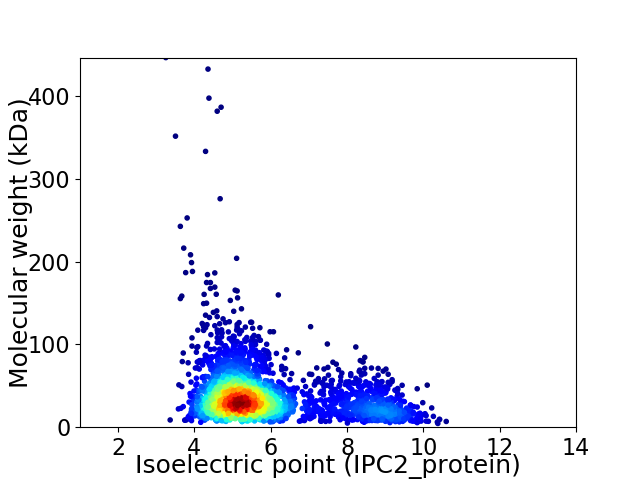
Virtual 2D-PAGE plot for 2884 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L3ZF07|A0A6L3ZF07_9FLAO TolC family protein OS=Phaeocystidibacter marisrubri OX=1577780 GN=F8C82_09895 PE=3 SV=1
MM1 pKa = 7.55KK2 pKa = 10.47KK3 pKa = 10.35FLFFLLASLLNLSTFQSIAQGVNFEE28 pKa = 4.68LMGSFGPHH36 pKa = 4.51EE37 pKa = 4.13QVKK40 pKa = 9.88VEE42 pKa = 4.68LDD44 pKa = 3.3QLYY47 pKa = 11.03DD48 pKa = 3.6PMSDD52 pKa = 3.04IGDD55 pKa = 3.46VDD57 pKa = 3.74GDD59 pKa = 3.86GDD61 pKa = 4.73LDD63 pKa = 4.03IIIAGEE69 pKa = 4.03TGSTSRR75 pKa = 11.84ITKK78 pKa = 9.28IYY80 pKa = 9.7LYY82 pKa = 10.2NGSDD86 pKa = 3.76SYY88 pKa = 12.09DD89 pKa = 3.41SLAGIGIIGVLPRR102 pKa = 11.84ALRR105 pKa = 11.84LIDD108 pKa = 4.84LDD110 pKa = 4.09GDD112 pKa = 3.71SDD114 pKa = 5.18LDD116 pKa = 3.75VFIAGITSSNQSVTRR131 pKa = 11.84IYY133 pKa = 10.91TNDD136 pKa = 3.12GTGGFTQISSSQLPSDD152 pKa = 2.84IWSAEE157 pKa = 3.76FGDD160 pKa = 4.65LDD162 pKa = 4.14NDD164 pKa = 3.95GDD166 pKa = 3.91VDD168 pKa = 4.56LVLSTSSFNVVIFDD182 pKa = 3.78NDD184 pKa = 3.53SNASFSLRR192 pKa = 11.84SGLSLPQLKK201 pKa = 10.58ADD203 pKa = 5.13DD204 pKa = 4.73IEE206 pKa = 4.52LADD209 pKa = 3.59INYY212 pKa = 10.4DD213 pKa = 3.58SFIDD217 pKa = 3.4IVYY220 pKa = 10.27AGSVPGGAAGQLHH233 pKa = 6.57AVYY236 pKa = 10.31NDD238 pKa = 3.65SNSGFTSYY246 pKa = 11.44LLNQASFLPNGGIKK260 pKa = 9.68CANYY264 pKa = 10.67NGDD267 pKa = 3.61SLMDD271 pKa = 3.43ILYY274 pKa = 10.95VGDD277 pKa = 3.99AVSGLLLLQDD287 pKa = 3.59STGVFSLKK295 pKa = 10.85SNIYY299 pKa = 8.35PQGVLNPTIYY309 pKa = 10.53AYY311 pKa = 10.59DD312 pKa = 3.78RR313 pKa = 11.84DD314 pKa = 3.96GDD316 pKa = 3.81GDD318 pKa = 4.08EE319 pKa = 5.69DD320 pKa = 4.15ILVLGVNRR328 pKa = 11.84SGVTVYY334 pKa = 8.85HH335 pKa = 6.36TFYY338 pKa = 11.23DD339 pKa = 3.64NYY341 pKa = 10.72GDD343 pKa = 4.22GSFSPVSGHH352 pKa = 6.57GLDD355 pKa = 3.77ARR357 pKa = 11.84LPNFTPRR364 pKa = 11.84YY365 pKa = 8.86FSSQLADD372 pKa = 3.77FNGDD376 pKa = 3.39GEE378 pKa = 4.58LDD380 pKa = 3.3ILGGRR385 pKa = 11.84DD386 pKa = 3.22PGVWDD391 pKa = 5.08LYY393 pKa = 10.57HH394 pKa = 6.28GHH396 pKa = 6.55SNGKK400 pKa = 9.49FYY402 pKa = 10.7HH403 pKa = 6.78ASLSSFGNITAQNQAVADD421 pKa = 4.37FNSDD425 pKa = 3.06GALDD429 pKa = 3.74IVVVNYY435 pKa = 10.69DD436 pKa = 3.18STRR439 pKa = 11.84VYY441 pKa = 10.64FNSGTGTFPTSKK453 pKa = 10.67GLDD456 pKa = 3.14HH457 pKa = 7.33FSYY460 pKa = 10.88YY461 pKa = 9.83WGDD464 pKa = 3.18IATGDD469 pKa = 3.81FDD471 pKa = 5.38NDD473 pKa = 3.2GDD475 pKa = 3.96EE476 pKa = 5.77DD477 pKa = 4.17IVFSGSFDD485 pKa = 3.32VTKK488 pKa = 10.64IFWNNGSGDD497 pKa = 3.8FSSAPTLFTEE507 pKa = 5.28MFQQANIEE515 pKa = 4.28VFDD518 pKa = 3.9VDD520 pKa = 4.33NDD522 pKa = 3.58ADD524 pKa = 4.51LDD526 pKa = 3.92ILVTGASNSSYY537 pKa = 11.11EE538 pKa = 3.97NTALYY543 pKa = 10.1INNGSGQFVVKK554 pKa = 9.95QNSGIPPTWGSTSAQGDD571 pKa = 3.29IDD573 pKa = 4.97LDD575 pKa = 3.94GDD577 pKa = 3.61IDD579 pKa = 4.36LVLTGSPGSSIYY591 pKa = 11.06LNDD594 pKa = 3.7GQGVFTTAPMPFSTNFIMPFCEE616 pKa = 4.27LVDD619 pKa = 4.15FNGDD623 pKa = 3.56SLPDD627 pKa = 3.4MFASRR632 pKa = 11.84GSTPDD637 pKa = 2.84ATIFEE642 pKa = 4.41NLGDD646 pKa = 3.58GSFAYY651 pKa = 8.35RR652 pKa = 11.84TRR654 pKa = 11.84FGLLTQMIICDD665 pKa = 3.91ANHH668 pKa = 7.26DD669 pKa = 4.28GFPDD673 pKa = 3.26ILTGGKK679 pKa = 10.03VYY681 pKa = 11.2VNDD684 pKa = 4.15GPGNFTFTLEE694 pKa = 4.3SNTGIRR700 pKa = 11.84TEE702 pKa = 4.02NSSGGLGIAADD713 pKa = 4.2FDD715 pKa = 4.77GDD717 pKa = 3.55TDD719 pKa = 5.05DD720 pKa = 6.18DD721 pKa = 3.72IFYY724 pKa = 10.73ASAYY728 pKa = 9.12GYY730 pKa = 10.18QGNFYY735 pKa = 10.78LRR737 pKa = 11.84YY738 pKa = 6.82TQLYY742 pKa = 8.73RR743 pKa = 11.84NMAVGLSVVEE753 pKa = 4.08NTSRR757 pKa = 11.84QQGISPYY764 pKa = 8.37PNPMVSDD771 pKa = 3.59ATVDD775 pKa = 3.73LPEE778 pKa = 4.62GTFSGQIIEE787 pKa = 4.08VSGRR791 pKa = 11.84IVRR794 pKa = 11.84QINNASGNLVIEE806 pKa = 4.22RR807 pKa = 11.84EE808 pKa = 4.18GLNIGAYY815 pKa = 9.74VLVLTGEE822 pKa = 4.26NGEE825 pKa = 4.35SFTTKK830 pKa = 10.47FLVQQ834 pKa = 3.47
MM1 pKa = 7.55KK2 pKa = 10.47KK3 pKa = 10.35FLFFLLASLLNLSTFQSIAQGVNFEE28 pKa = 4.68LMGSFGPHH36 pKa = 4.51EE37 pKa = 4.13QVKK40 pKa = 9.88VEE42 pKa = 4.68LDD44 pKa = 3.3QLYY47 pKa = 11.03DD48 pKa = 3.6PMSDD52 pKa = 3.04IGDD55 pKa = 3.46VDD57 pKa = 3.74GDD59 pKa = 3.86GDD61 pKa = 4.73LDD63 pKa = 4.03IIIAGEE69 pKa = 4.03TGSTSRR75 pKa = 11.84ITKK78 pKa = 9.28IYY80 pKa = 9.7LYY82 pKa = 10.2NGSDD86 pKa = 3.76SYY88 pKa = 12.09DD89 pKa = 3.41SLAGIGIIGVLPRR102 pKa = 11.84ALRR105 pKa = 11.84LIDD108 pKa = 4.84LDD110 pKa = 4.09GDD112 pKa = 3.71SDD114 pKa = 5.18LDD116 pKa = 3.75VFIAGITSSNQSVTRR131 pKa = 11.84IYY133 pKa = 10.91TNDD136 pKa = 3.12GTGGFTQISSSQLPSDD152 pKa = 2.84IWSAEE157 pKa = 3.76FGDD160 pKa = 4.65LDD162 pKa = 4.14NDD164 pKa = 3.95GDD166 pKa = 3.91VDD168 pKa = 4.56LVLSTSSFNVVIFDD182 pKa = 3.78NDD184 pKa = 3.53SNASFSLRR192 pKa = 11.84SGLSLPQLKK201 pKa = 10.58ADD203 pKa = 5.13DD204 pKa = 4.73IEE206 pKa = 4.52LADD209 pKa = 3.59INYY212 pKa = 10.4DD213 pKa = 3.58SFIDD217 pKa = 3.4IVYY220 pKa = 10.27AGSVPGGAAGQLHH233 pKa = 6.57AVYY236 pKa = 10.31NDD238 pKa = 3.65SNSGFTSYY246 pKa = 11.44LLNQASFLPNGGIKK260 pKa = 9.68CANYY264 pKa = 10.67NGDD267 pKa = 3.61SLMDD271 pKa = 3.43ILYY274 pKa = 10.95VGDD277 pKa = 3.99AVSGLLLLQDD287 pKa = 3.59STGVFSLKK295 pKa = 10.85SNIYY299 pKa = 8.35PQGVLNPTIYY309 pKa = 10.53AYY311 pKa = 10.59DD312 pKa = 3.78RR313 pKa = 11.84DD314 pKa = 3.96GDD316 pKa = 3.81GDD318 pKa = 4.08EE319 pKa = 5.69DD320 pKa = 4.15ILVLGVNRR328 pKa = 11.84SGVTVYY334 pKa = 8.85HH335 pKa = 6.36TFYY338 pKa = 11.23DD339 pKa = 3.64NYY341 pKa = 10.72GDD343 pKa = 4.22GSFSPVSGHH352 pKa = 6.57GLDD355 pKa = 3.77ARR357 pKa = 11.84LPNFTPRR364 pKa = 11.84YY365 pKa = 8.86FSSQLADD372 pKa = 3.77FNGDD376 pKa = 3.39GEE378 pKa = 4.58LDD380 pKa = 3.3ILGGRR385 pKa = 11.84DD386 pKa = 3.22PGVWDD391 pKa = 5.08LYY393 pKa = 10.57HH394 pKa = 6.28GHH396 pKa = 6.55SNGKK400 pKa = 9.49FYY402 pKa = 10.7HH403 pKa = 6.78ASLSSFGNITAQNQAVADD421 pKa = 4.37FNSDD425 pKa = 3.06GALDD429 pKa = 3.74IVVVNYY435 pKa = 10.69DD436 pKa = 3.18STRR439 pKa = 11.84VYY441 pKa = 10.64FNSGTGTFPTSKK453 pKa = 10.67GLDD456 pKa = 3.14HH457 pKa = 7.33FSYY460 pKa = 10.88YY461 pKa = 9.83WGDD464 pKa = 3.18IATGDD469 pKa = 3.81FDD471 pKa = 5.38NDD473 pKa = 3.2GDD475 pKa = 3.96EE476 pKa = 5.77DD477 pKa = 4.17IVFSGSFDD485 pKa = 3.32VTKK488 pKa = 10.64IFWNNGSGDD497 pKa = 3.8FSSAPTLFTEE507 pKa = 5.28MFQQANIEE515 pKa = 4.28VFDD518 pKa = 3.9VDD520 pKa = 4.33NDD522 pKa = 3.58ADD524 pKa = 4.51LDD526 pKa = 3.92ILVTGASNSSYY537 pKa = 11.11EE538 pKa = 3.97NTALYY543 pKa = 10.1INNGSGQFVVKK554 pKa = 9.95QNSGIPPTWGSTSAQGDD571 pKa = 3.29IDD573 pKa = 4.97LDD575 pKa = 3.94GDD577 pKa = 3.61IDD579 pKa = 4.36LVLTGSPGSSIYY591 pKa = 11.06LNDD594 pKa = 3.7GQGVFTTAPMPFSTNFIMPFCEE616 pKa = 4.27LVDD619 pKa = 4.15FNGDD623 pKa = 3.56SLPDD627 pKa = 3.4MFASRR632 pKa = 11.84GSTPDD637 pKa = 2.84ATIFEE642 pKa = 4.41NLGDD646 pKa = 3.58GSFAYY651 pKa = 8.35RR652 pKa = 11.84TRR654 pKa = 11.84FGLLTQMIICDD665 pKa = 3.91ANHH668 pKa = 7.26DD669 pKa = 4.28GFPDD673 pKa = 3.26ILTGGKK679 pKa = 10.03VYY681 pKa = 11.2VNDD684 pKa = 4.15GPGNFTFTLEE694 pKa = 4.3SNTGIRR700 pKa = 11.84TEE702 pKa = 4.02NSSGGLGIAADD713 pKa = 4.2FDD715 pKa = 4.77GDD717 pKa = 3.55TDD719 pKa = 5.05DD720 pKa = 6.18DD721 pKa = 3.72IFYY724 pKa = 10.73ASAYY728 pKa = 9.12GYY730 pKa = 10.18QGNFYY735 pKa = 10.78LRR737 pKa = 11.84YY738 pKa = 6.82TQLYY742 pKa = 8.73RR743 pKa = 11.84NMAVGLSVVEE753 pKa = 4.08NTSRR757 pKa = 11.84QQGISPYY764 pKa = 8.37PNPMVSDD771 pKa = 3.59ATVDD775 pKa = 3.73LPEE778 pKa = 4.62GTFSGQIIEE787 pKa = 4.08VSGRR791 pKa = 11.84IVRR794 pKa = 11.84QINNASGNLVIEE806 pKa = 4.22RR807 pKa = 11.84EE808 pKa = 4.18GLNIGAYY815 pKa = 9.74VLVLTGEE822 pKa = 4.26NGEE825 pKa = 4.35SFTTKK830 pKa = 10.47FLVQQ834 pKa = 3.47
Molecular weight: 89.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L3ZD93|A0A6L3ZD93_9FLAO Uncharacterized protein OS=Phaeocystidibacter marisrubri OX=1577780 GN=F8C82_07895 PE=4 SV=1
MM1 pKa = 7.0NQTRR5 pKa = 11.84QKK7 pKa = 10.52RR8 pKa = 11.84LLRR11 pKa = 11.84TFEE14 pKa = 4.37FLWMIAAAVMVVEE27 pKa = 5.11CYY29 pKa = 10.27VAFSGGDD36 pKa = 3.35TTRR39 pKa = 11.84GLILLLLGLYY49 pKa = 10.53AGFRR53 pKa = 11.84YY54 pKa = 8.33VTTGMKK60 pKa = 9.68RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.35LL64 pKa = 3.54
MM1 pKa = 7.0NQTRR5 pKa = 11.84QKK7 pKa = 10.52RR8 pKa = 11.84LLRR11 pKa = 11.84TFEE14 pKa = 4.37FLWMIAAAVMVVEE27 pKa = 5.11CYY29 pKa = 10.27VAFSGGDD36 pKa = 3.35TTRR39 pKa = 11.84GLILLLLGLYY49 pKa = 10.53AGFRR53 pKa = 11.84YY54 pKa = 8.33VTTGMKK60 pKa = 9.68RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.35LL64 pKa = 3.54
Molecular weight: 7.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015842 |
38 |
4451 |
352.2 |
39.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.21 ± 0.046 | 0.767 ± 0.018 |
5.772 ± 0.033 | 6.639 ± 0.064 |
4.675 ± 0.037 | 7.192 ± 0.051 |
2.115 ± 0.021 | 6.579 ± 0.035 |
5.001 ± 0.057 | 9.168 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.023 | 4.947 ± 0.037 |
3.937 ± 0.032 | 3.419 ± 0.026 |
4.616 ± 0.045 | 7.227 ± 0.051 |
5.898 ± 0.071 | 6.956 ± 0.038 |
1.371 ± 0.019 | 3.984 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
