
Torque teno midi virus 12
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
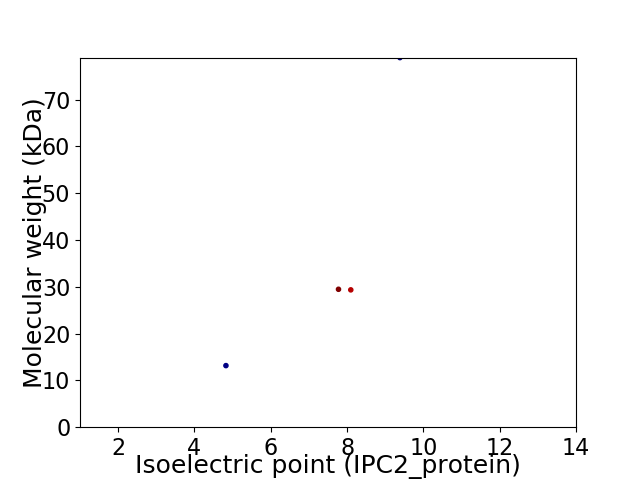
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7VLZ6|A7VLZ6_9VIRU Capsid protein OS=Torque teno midi virus 12 OX=2065053 PE=3 SV=1
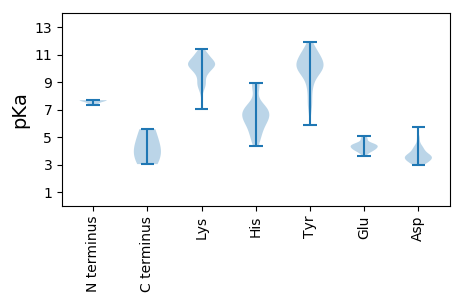
MM1 pKa = 7.68SIFYY5 pKa = 10.48KK6 pKa = 8.57PAKK9 pKa = 10.01YY10 pKa = 9.94NGHH13 pKa = 6.35TKK15 pKa = 8.88QQMWMSVIADD25 pKa = 3.81FHH27 pKa = 8.93DD28 pKa = 4.2PMCNCWHH35 pKa = 7.12PFGHH39 pKa = 7.09LLDD42 pKa = 4.93IIFPEE47 pKa = 4.28GHH49 pKa = 7.13KK50 pKa = 11.1DD51 pKa = 3.3RR52 pKa = 11.84DD53 pKa = 3.46KK54 pKa = 11.21TIAQIITRR62 pKa = 11.84DD63 pKa = 4.07SKK65 pKa = 10.92CHH67 pKa = 6.45SGGEE71 pKa = 4.18DD72 pKa = 3.11EE73 pKa = 4.96GASGQTEE80 pKa = 3.97DD81 pKa = 4.38LHH83 pKa = 8.65IKK85 pKa = 8.78EE86 pKa = 4.85EE87 pKa = 4.43RR88 pKa = 11.84DD89 pKa = 3.55TPADD93 pKa = 3.6TTGGMAEE100 pKa = 4.51EE101 pKa = 4.45EE102 pKa = 4.36TTDD105 pKa = 3.58ALLAAAVEE113 pKa = 4.28NFEE116 pKa = 4.35RR117 pKa = 5.57
MM1 pKa = 7.68SIFYY5 pKa = 10.48KK6 pKa = 8.57PAKK9 pKa = 10.01YY10 pKa = 9.94NGHH13 pKa = 6.35TKK15 pKa = 8.88QQMWMSVIADD25 pKa = 3.81FHH27 pKa = 8.93DD28 pKa = 4.2PMCNCWHH35 pKa = 7.12PFGHH39 pKa = 7.09LLDD42 pKa = 4.93IIFPEE47 pKa = 4.28GHH49 pKa = 7.13KK50 pKa = 11.1DD51 pKa = 3.3RR52 pKa = 11.84DD53 pKa = 3.46KK54 pKa = 11.21TIAQIITRR62 pKa = 11.84DD63 pKa = 4.07SKK65 pKa = 10.92CHH67 pKa = 6.45SGGEE71 pKa = 4.18DD72 pKa = 3.11EE73 pKa = 4.96GASGQTEE80 pKa = 3.97DD81 pKa = 4.38LHH83 pKa = 8.65IKK85 pKa = 8.78EE86 pKa = 4.85EE87 pKa = 4.43RR88 pKa = 11.84DD89 pKa = 3.55TPADD93 pKa = 3.6TTGGMAEE100 pKa = 4.51EE101 pKa = 4.45EE102 pKa = 4.36TTDD105 pKa = 3.58ALLAAAVEE113 pKa = 4.28NFEE116 pKa = 4.35RR117 pKa = 5.57
Molecular weight: 13.17 kDa
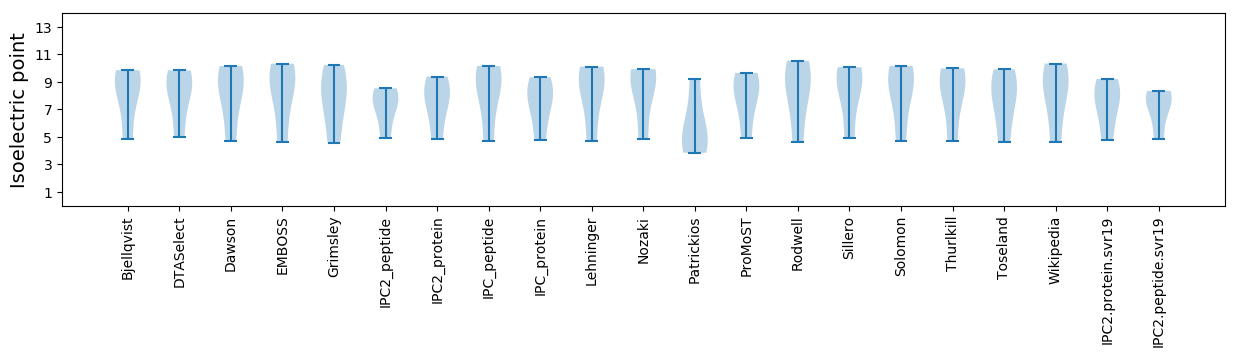
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLZ6|A7VLZ6_9VIRU Capsid protein OS=Torque teno midi virus 12 OX=2065053 PE=3 SV=1
MM1 pKa = 7.33PFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84FWTNRR15 pKa = 11.84RR16 pKa = 11.84FTYY19 pKa = 9.93KK20 pKa = 9.74RR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.45RR24 pKa = 11.84YY25 pKa = 7.52PRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 7.39NRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.35GRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NYY39 pKa = 9.67RR40 pKa = 11.84RR41 pKa = 11.84PARR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.07LRR51 pKa = 11.84KK52 pKa = 8.85VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.67KK57 pKa = 10.41KK58 pKa = 10.3QIAVKK63 pKa = 9.38QWQPEE68 pKa = 4.47SITKK72 pKa = 10.26CKK74 pKa = 10.25ILGHH78 pKa = 5.77SALVVGAEE86 pKa = 4.34GKK88 pKa = 10.5QMFCYY93 pKa = 7.45TTNKK97 pKa = 9.88NIAVPPKK104 pKa = 9.63SPYY107 pKa = 9.86GGGFGVEE114 pKa = 4.17QYY116 pKa = 11.19SLNYY120 pKa = 10.23LYY122 pKa = 10.89EE123 pKa = 4.41EE124 pKa = 4.54YY125 pKa = 10.22TFHH128 pKa = 6.8NNYY131 pKa = 5.91WTKK134 pKa = 11.36SNILKK139 pKa = 10.08DD140 pKa = 3.35LCRR143 pKa = 11.84FLWLKK148 pKa = 10.73IIFFRR153 pKa = 11.84DD154 pKa = 2.96NKK156 pKa = 9.83TDD158 pKa = 4.52FIVSYY163 pKa = 10.83DD164 pKa = 3.64RR165 pKa = 11.84NPPFNLTKK173 pKa = 9.64FTYY176 pKa = 10.04PGAHH180 pKa = 6.39PQQMLLQKK188 pKa = 9.62HH189 pKa = 6.17HH190 pKa = 6.91KK191 pKa = 10.16LILSQMTKK199 pKa = 10.66NPNKK203 pKa = 9.05LTKK206 pKa = 9.85KK207 pKa = 10.45FKK209 pKa = 10.38IKK211 pKa = 10.18PPRR214 pKa = 11.84QMISKK219 pKa = 9.26WFFTKK224 pKa = 10.37PFAKK228 pKa = 10.56YY229 pKa = 9.72PLLSLKK235 pKa = 10.68ASALDD240 pKa = 3.81LRR242 pKa = 11.84HH243 pKa = 6.74SYY245 pKa = 10.5LGCCNEE251 pKa = 3.61NSQVYY256 pKa = 8.4FFYY259 pKa = 10.85LNHH262 pKa = 6.54GFYY265 pKa = 10.57KK266 pKa = 8.23QTNWGQYY273 pKa = 7.05STTPYY278 pKa = 10.25RR279 pKa = 11.84PSNEE283 pKa = 3.82INTVYY288 pKa = 11.06YY289 pKa = 10.32KK290 pKa = 11.0DD291 pKa = 3.09KK292 pKa = 10.8TGNKK296 pKa = 8.79KK297 pKa = 10.47QIGPFNTTATAYY309 pKa = 9.95DD310 pKa = 3.89DD311 pKa = 3.83SVSYY315 pKa = 9.97TKK317 pKa = 10.68GYY319 pKa = 8.44FQPPFLQTQFVCTSSMCDD337 pKa = 3.01VKK339 pKa = 11.06AGYY342 pKa = 10.23LPINVAIYY350 pKa = 11.17NPMQDD355 pKa = 3.11SSQGNAIYY363 pKa = 10.29LVSTLTNTWDD373 pKa = 3.6QPPKK377 pKa = 10.47DD378 pKa = 4.08SSILIQGMPIWLGLFGYY395 pKa = 10.32LDD397 pKa = 3.72YY398 pKa = 11.35VRR400 pKa = 11.84QVKK403 pKa = 9.87KK404 pKa = 10.81DD405 pKa = 3.65KK406 pKa = 9.98TWLEE410 pKa = 4.0SHH412 pKa = 6.26VLVLKK417 pKa = 10.87SPAIFTFPQPGADD430 pKa = 2.96AWYY433 pKa = 9.96CPLSKK438 pKa = 9.85TFIDD442 pKa = 4.24GVGPYY447 pKa = 8.78RR448 pKa = 11.84TDD450 pKa = 3.09LTIKK454 pKa = 10.3QKK456 pKa = 10.2EE457 pKa = 3.89RR458 pKa = 11.84WFPQIQHH465 pKa = 5.67QYY467 pKa = 9.72EE468 pKa = 4.07ILNSFVEE475 pKa = 4.63AGPFIPKK482 pKa = 8.38YY483 pKa = 8.7TNQSEE488 pKa = 5.07SNWEE492 pKa = 3.97LKK494 pKa = 10.18FKK496 pKa = 11.14YY497 pKa = 9.58IFHH500 pKa = 6.81FKK502 pKa = 9.76WGGPSLNEE510 pKa = 3.79PDD512 pKa = 3.13ITNPANQDD520 pKa = 3.21QYY522 pKa = 11.93DD523 pKa = 3.91VPDD526 pKa = 3.98TLQQTIQIEE535 pKa = 4.27NPEE538 pKa = 4.15RR539 pKa = 11.84QDD541 pKa = 3.14PRR543 pKa = 11.84SILHH547 pKa = 4.78QWDD550 pKa = 3.39YY551 pKa = 11.42RR552 pKa = 11.84RR553 pKa = 11.84GFIKK557 pKa = 10.29EE558 pKa = 3.72RR559 pKa = 11.84ALKK562 pKa = 10.64RR563 pKa = 11.84MSTYY567 pKa = 10.85LSTPSDD573 pKa = 3.62VPEE576 pKa = 4.62SPEE579 pKa = 3.68HH580 pKa = 5.31TPKK583 pKa = 10.39KK584 pKa = 9.17KK585 pKa = 10.31KK586 pKa = 10.13RR587 pKa = 11.84VGAQLTLPQEE597 pKa = 4.52KK598 pKa = 10.39DD599 pKa = 3.32QEE601 pKa = 4.52TLSCLLSLCKK611 pKa = 10.11EE612 pKa = 4.82DD613 pKa = 5.7IYY615 pKa = 11.24QKK617 pKa = 10.39PQTEE621 pKa = 4.2EE622 pKa = 4.32EE623 pKa = 4.22INQLIQQQQQQQHH636 pKa = 4.32QLKK639 pKa = 10.26RR640 pKa = 11.84DD641 pKa = 3.45ILHH644 pKa = 6.82LMMQIRR650 pKa = 11.84EE651 pKa = 4.1QQQLLQLHH659 pKa = 6.16TGIIPP664 pKa = 3.81
MM1 pKa = 7.33PFWWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84FWTNRR15 pKa = 11.84RR16 pKa = 11.84FTYY19 pKa = 9.93KK20 pKa = 9.74RR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.45RR24 pKa = 11.84YY25 pKa = 7.52PRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 7.39NRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.35GRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NYY39 pKa = 9.67RR40 pKa = 11.84RR41 pKa = 11.84PARR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.07LRR51 pKa = 11.84KK52 pKa = 8.85VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.67KK57 pKa = 10.41KK58 pKa = 10.3QIAVKK63 pKa = 9.38QWQPEE68 pKa = 4.47SITKK72 pKa = 10.26CKK74 pKa = 10.25ILGHH78 pKa = 5.77SALVVGAEE86 pKa = 4.34GKK88 pKa = 10.5QMFCYY93 pKa = 7.45TTNKK97 pKa = 9.88NIAVPPKK104 pKa = 9.63SPYY107 pKa = 9.86GGGFGVEE114 pKa = 4.17QYY116 pKa = 11.19SLNYY120 pKa = 10.23LYY122 pKa = 10.89EE123 pKa = 4.41EE124 pKa = 4.54YY125 pKa = 10.22TFHH128 pKa = 6.8NNYY131 pKa = 5.91WTKK134 pKa = 11.36SNILKK139 pKa = 10.08DD140 pKa = 3.35LCRR143 pKa = 11.84FLWLKK148 pKa = 10.73IIFFRR153 pKa = 11.84DD154 pKa = 2.96NKK156 pKa = 9.83TDD158 pKa = 4.52FIVSYY163 pKa = 10.83DD164 pKa = 3.64RR165 pKa = 11.84NPPFNLTKK173 pKa = 9.64FTYY176 pKa = 10.04PGAHH180 pKa = 6.39PQQMLLQKK188 pKa = 9.62HH189 pKa = 6.17HH190 pKa = 6.91KK191 pKa = 10.16LILSQMTKK199 pKa = 10.66NPNKK203 pKa = 9.05LTKK206 pKa = 9.85KK207 pKa = 10.45FKK209 pKa = 10.38IKK211 pKa = 10.18PPRR214 pKa = 11.84QMISKK219 pKa = 9.26WFFTKK224 pKa = 10.37PFAKK228 pKa = 10.56YY229 pKa = 9.72PLLSLKK235 pKa = 10.68ASALDD240 pKa = 3.81LRR242 pKa = 11.84HH243 pKa = 6.74SYY245 pKa = 10.5LGCCNEE251 pKa = 3.61NSQVYY256 pKa = 8.4FFYY259 pKa = 10.85LNHH262 pKa = 6.54GFYY265 pKa = 10.57KK266 pKa = 8.23QTNWGQYY273 pKa = 7.05STTPYY278 pKa = 10.25RR279 pKa = 11.84PSNEE283 pKa = 3.82INTVYY288 pKa = 11.06YY289 pKa = 10.32KK290 pKa = 11.0DD291 pKa = 3.09KK292 pKa = 10.8TGNKK296 pKa = 8.79KK297 pKa = 10.47QIGPFNTTATAYY309 pKa = 9.95DD310 pKa = 3.89DD311 pKa = 3.83SVSYY315 pKa = 9.97TKK317 pKa = 10.68GYY319 pKa = 8.44FQPPFLQTQFVCTSSMCDD337 pKa = 3.01VKK339 pKa = 11.06AGYY342 pKa = 10.23LPINVAIYY350 pKa = 11.17NPMQDD355 pKa = 3.11SSQGNAIYY363 pKa = 10.29LVSTLTNTWDD373 pKa = 3.6QPPKK377 pKa = 10.47DD378 pKa = 4.08SSILIQGMPIWLGLFGYY395 pKa = 10.32LDD397 pKa = 3.72YY398 pKa = 11.35VRR400 pKa = 11.84QVKK403 pKa = 9.87KK404 pKa = 10.81DD405 pKa = 3.65KK406 pKa = 9.98TWLEE410 pKa = 4.0SHH412 pKa = 6.26VLVLKK417 pKa = 10.87SPAIFTFPQPGADD430 pKa = 2.96AWYY433 pKa = 9.96CPLSKK438 pKa = 9.85TFIDD442 pKa = 4.24GVGPYY447 pKa = 8.78RR448 pKa = 11.84TDD450 pKa = 3.09LTIKK454 pKa = 10.3QKK456 pKa = 10.2EE457 pKa = 3.89RR458 pKa = 11.84WFPQIQHH465 pKa = 5.67QYY467 pKa = 9.72EE468 pKa = 4.07ILNSFVEE475 pKa = 4.63AGPFIPKK482 pKa = 8.38YY483 pKa = 8.7TNQSEE488 pKa = 5.07SNWEE492 pKa = 3.97LKK494 pKa = 10.18FKK496 pKa = 11.14YY497 pKa = 9.58IFHH500 pKa = 6.81FKK502 pKa = 9.76WGGPSLNEE510 pKa = 3.79PDD512 pKa = 3.13ITNPANQDD520 pKa = 3.21QYY522 pKa = 11.93DD523 pKa = 3.91VPDD526 pKa = 3.98TLQQTIQIEE535 pKa = 4.27NPEE538 pKa = 4.15RR539 pKa = 11.84QDD541 pKa = 3.14PRR543 pKa = 11.84SILHH547 pKa = 4.78QWDD550 pKa = 3.39YY551 pKa = 11.42RR552 pKa = 11.84RR553 pKa = 11.84GFIKK557 pKa = 10.29EE558 pKa = 3.72RR559 pKa = 11.84ALKK562 pKa = 10.64RR563 pKa = 11.84MSTYY567 pKa = 10.85LSTPSDD573 pKa = 3.62VPEE576 pKa = 4.62SPEE579 pKa = 3.68HH580 pKa = 5.31TPKK583 pKa = 10.39KK584 pKa = 9.17KK585 pKa = 10.31KK586 pKa = 10.13RR587 pKa = 11.84VGAQLTLPQEE597 pKa = 4.52KK598 pKa = 10.39DD599 pKa = 3.32QEE601 pKa = 4.52TLSCLLSLCKK611 pKa = 10.11EE612 pKa = 4.82DD613 pKa = 5.7IYY615 pKa = 11.24QKK617 pKa = 10.39PQTEE621 pKa = 4.2EE622 pKa = 4.32EE623 pKa = 4.22INQLIQQQQQQQHH636 pKa = 4.32QLKK639 pKa = 10.26RR640 pKa = 11.84DD641 pKa = 3.45ILHH644 pKa = 6.82LMMQIRR650 pKa = 11.84EE651 pKa = 4.1QQQLLQLHH659 pKa = 6.16TGIIPP664 pKa = 3.81
Molecular weight: 79.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1300 |
117 |
664 |
325.0 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.154 ± 1.365 | 1.692 ± 0.19 |
5.385 ± 0.966 | 5.615 ± 1.175 |
4.385 ± 0.632 | 4.769 ± 0.472 |
3.923 ± 1.096 | 5.462 ± 0.298 |
8.846 ± 0.474 | 7.077 ± 0.85 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.584 | 4.077 ± 0.69 |
6.154 ± 1.151 | 6.385 ± 1.17 |
5.923 ± 0.873 | 6.692 ± 1.537 |
8.077 ± 1.103 | 2.385 ± 0.696 |
1.846 ± 0.365 | 3.846 ± 1.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
