
Beihai barnacle virus 8
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Artoviridae; Hexartovirus; Barnacle hexartovirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
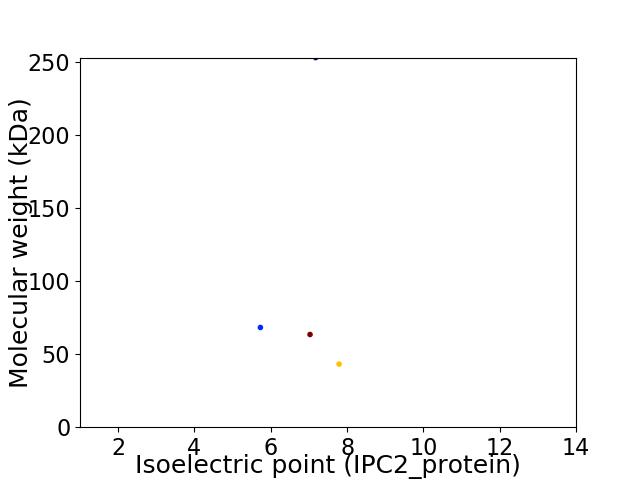
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMV2|A0A1L3KMV2_9MONO GDP polyribonucleotidyltransferase OS=Beihai barnacle virus 8 OX=1922366 PE=4 SV=1
MM1 pKa = 7.58ANADD5 pKa = 3.88PGDD8 pKa = 3.59EE9 pKa = 3.91RR10 pKa = 11.84LRR12 pKa = 11.84ALEE15 pKa = 4.06TFSSILHH22 pKa = 5.96HH23 pKa = 6.68LGIEE27 pKa = 4.12AEE29 pKa = 4.03EE30 pKa = 4.34SNRR33 pKa = 11.84GFVQLGTVQHH43 pKa = 5.72VSYY46 pKa = 11.01LFGKK50 pKa = 7.28QTEE53 pKa = 4.15PAIVHH58 pKa = 5.14TTSDD62 pKa = 3.83LLRR65 pKa = 11.84QTNEE69 pKa = 3.66FAPDD73 pKa = 2.98EE74 pKa = 4.43GFFLSGMLTRR84 pKa = 11.84GGKK87 pKa = 9.73SYY89 pKa = 10.76TSLYY93 pKa = 9.83VIQDD97 pKa = 3.54KK98 pKa = 11.09VLEE101 pKa = 4.4DD102 pKa = 3.52QGLLLNIQRR111 pKa = 11.84RR112 pKa = 11.84KK113 pKa = 10.04EE114 pKa = 3.89LLDD117 pKa = 3.48MSEE120 pKa = 4.09MTLKK124 pKa = 10.59RR125 pKa = 11.84MKK127 pKa = 10.49QLGKK131 pKa = 8.95SAKK134 pKa = 9.93VVFQSDD140 pKa = 4.5VINSIPIHH148 pKa = 5.29LVKK151 pKa = 10.65PEE153 pKa = 4.46LLPEE157 pKa = 4.2IAADD161 pKa = 3.63NRR163 pKa = 11.84LSRR166 pKa = 11.84ASKK169 pKa = 9.55VASALFSVFPWTLDD183 pKa = 3.12NASAVFHH190 pKa = 6.0AQQYY194 pKa = 10.21DD195 pKa = 3.61KK196 pKa = 10.9EE197 pKa = 4.27ASALEE202 pKa = 4.22LPPGVNAQSEE212 pKa = 4.3EE213 pKa = 4.12GAAQLDD219 pKa = 3.92TALRR223 pKa = 11.84NQFHH227 pKa = 6.01QMVIAVGTLCLCYY240 pKa = 10.2GYY242 pKa = 10.6SQDD245 pKa = 4.9NLIAKK250 pKa = 8.9QISRR254 pKa = 11.84RR255 pKa = 11.84LAAAYY260 pKa = 10.43ADD262 pKa = 4.36ANLAAPDD269 pKa = 3.79WDD271 pKa = 3.73TVANNIKK278 pKa = 9.46EE279 pKa = 4.34WAGSPRR285 pKa = 11.84EE286 pKa = 3.78IHH288 pKa = 6.34GEE290 pKa = 4.08VVALIGRR297 pKa = 11.84VLALHH302 pKa = 7.19RR303 pKa = 11.84DD304 pKa = 3.63GSINPSLAAEE314 pKa = 4.5DD315 pKa = 4.07EE316 pKa = 4.39ALSAEE321 pKa = 4.32DD322 pKa = 3.46PRR324 pKa = 11.84LRR326 pKa = 11.84NLPPIFNAVRR336 pKa = 11.84DD337 pKa = 3.95QMLLVYY343 pKa = 9.87TNYY346 pKa = 9.95QSSTIRR352 pKa = 11.84WALQVLPLVEE362 pKa = 3.92EE363 pKa = 4.58AGFGTHH369 pKa = 6.09HH370 pKa = 7.29RR371 pKa = 11.84LGAEE375 pKa = 3.49IRR377 pKa = 11.84HH378 pKa = 5.83LKK380 pKa = 9.38SIKK383 pKa = 10.72GEE385 pKa = 3.89IEE387 pKa = 3.51GRR389 pKa = 11.84KK390 pKa = 9.37YY391 pKa = 10.79VGLVSKK397 pKa = 10.56IATRR401 pKa = 11.84HH402 pKa = 4.29QQQQFPMMSFIGCMYY417 pKa = 10.56HH418 pKa = 7.8SMHH421 pKa = 7.32LEE423 pKa = 4.08TEE425 pKa = 4.16DD426 pKa = 2.94QKK428 pKa = 11.97AAFRR432 pKa = 11.84DD433 pKa = 4.34FNLTGISQKK442 pKa = 10.53VADD445 pKa = 4.18VQSKK449 pKa = 8.24PVCDD453 pKa = 4.33AVLKK457 pKa = 8.53MLPAPHH463 pKa = 6.83IEE465 pKa = 4.18AKK467 pKa = 10.31ASLMKK472 pKa = 10.64GMSALDD478 pKa = 3.57VEE480 pKa = 5.13RR481 pKa = 11.84VLSASPEE488 pKa = 4.17DD489 pKa = 3.82EE490 pKa = 4.12VKK492 pKa = 10.69LLKK495 pKa = 10.37EE496 pKa = 4.04RR497 pKa = 11.84LRR499 pKa = 11.84GVDD502 pKa = 4.81PPCDD506 pKa = 3.48WAIHH510 pKa = 5.52DD511 pKa = 4.64AKK513 pKa = 10.65EE514 pKa = 3.67QAAEE518 pKa = 4.06AKK520 pKa = 10.3KK521 pKa = 10.63KK522 pKa = 10.26FYY524 pKa = 10.34QAAALEE530 pKa = 4.58LNTRR534 pKa = 11.84WEE536 pKa = 4.21KK537 pKa = 11.12AHH539 pKa = 5.91ATMMDD544 pKa = 4.93RR545 pKa = 11.84ITMEE549 pKa = 4.22ANAEE553 pKa = 4.09TRR555 pKa = 11.84QRR557 pKa = 11.84LAQGIQTLRR566 pKa = 11.84AEE568 pKa = 4.64LLRR571 pKa = 11.84LTTAEE576 pKa = 4.13EE577 pKa = 4.3MSGSDD582 pKa = 5.26LMKK585 pKa = 10.37QLQITYY591 pKa = 9.97RR592 pKa = 11.84ADD594 pKa = 4.4LEE596 pKa = 4.46EE597 pKa = 4.08HH598 pKa = 6.38WKK600 pKa = 11.05AGMAVIQKK608 pKa = 9.92LRR610 pKa = 11.84EE611 pKa = 3.94LL612 pKa = 4.02
MM1 pKa = 7.58ANADD5 pKa = 3.88PGDD8 pKa = 3.59EE9 pKa = 3.91RR10 pKa = 11.84LRR12 pKa = 11.84ALEE15 pKa = 4.06TFSSILHH22 pKa = 5.96HH23 pKa = 6.68LGIEE27 pKa = 4.12AEE29 pKa = 4.03EE30 pKa = 4.34SNRR33 pKa = 11.84GFVQLGTVQHH43 pKa = 5.72VSYY46 pKa = 11.01LFGKK50 pKa = 7.28QTEE53 pKa = 4.15PAIVHH58 pKa = 5.14TTSDD62 pKa = 3.83LLRR65 pKa = 11.84QTNEE69 pKa = 3.66FAPDD73 pKa = 2.98EE74 pKa = 4.43GFFLSGMLTRR84 pKa = 11.84GGKK87 pKa = 9.73SYY89 pKa = 10.76TSLYY93 pKa = 9.83VIQDD97 pKa = 3.54KK98 pKa = 11.09VLEE101 pKa = 4.4DD102 pKa = 3.52QGLLLNIQRR111 pKa = 11.84RR112 pKa = 11.84KK113 pKa = 10.04EE114 pKa = 3.89LLDD117 pKa = 3.48MSEE120 pKa = 4.09MTLKK124 pKa = 10.59RR125 pKa = 11.84MKK127 pKa = 10.49QLGKK131 pKa = 8.95SAKK134 pKa = 9.93VVFQSDD140 pKa = 4.5VINSIPIHH148 pKa = 5.29LVKK151 pKa = 10.65PEE153 pKa = 4.46LLPEE157 pKa = 4.2IAADD161 pKa = 3.63NRR163 pKa = 11.84LSRR166 pKa = 11.84ASKK169 pKa = 9.55VASALFSVFPWTLDD183 pKa = 3.12NASAVFHH190 pKa = 6.0AQQYY194 pKa = 10.21DD195 pKa = 3.61KK196 pKa = 10.9EE197 pKa = 4.27ASALEE202 pKa = 4.22LPPGVNAQSEE212 pKa = 4.3EE213 pKa = 4.12GAAQLDD219 pKa = 3.92TALRR223 pKa = 11.84NQFHH227 pKa = 6.01QMVIAVGTLCLCYY240 pKa = 10.2GYY242 pKa = 10.6SQDD245 pKa = 4.9NLIAKK250 pKa = 8.9QISRR254 pKa = 11.84RR255 pKa = 11.84LAAAYY260 pKa = 10.43ADD262 pKa = 4.36ANLAAPDD269 pKa = 3.79WDD271 pKa = 3.73TVANNIKK278 pKa = 9.46EE279 pKa = 4.34WAGSPRR285 pKa = 11.84EE286 pKa = 3.78IHH288 pKa = 6.34GEE290 pKa = 4.08VVALIGRR297 pKa = 11.84VLALHH302 pKa = 7.19RR303 pKa = 11.84DD304 pKa = 3.63GSINPSLAAEE314 pKa = 4.5DD315 pKa = 4.07EE316 pKa = 4.39ALSAEE321 pKa = 4.32DD322 pKa = 3.46PRR324 pKa = 11.84LRR326 pKa = 11.84NLPPIFNAVRR336 pKa = 11.84DD337 pKa = 3.95QMLLVYY343 pKa = 9.87TNYY346 pKa = 9.95QSSTIRR352 pKa = 11.84WALQVLPLVEE362 pKa = 3.92EE363 pKa = 4.58AGFGTHH369 pKa = 6.09HH370 pKa = 7.29RR371 pKa = 11.84LGAEE375 pKa = 3.49IRR377 pKa = 11.84HH378 pKa = 5.83LKK380 pKa = 9.38SIKK383 pKa = 10.72GEE385 pKa = 3.89IEE387 pKa = 3.51GRR389 pKa = 11.84KK390 pKa = 9.37YY391 pKa = 10.79VGLVSKK397 pKa = 10.56IATRR401 pKa = 11.84HH402 pKa = 4.29QQQQFPMMSFIGCMYY417 pKa = 10.56HH418 pKa = 7.8SMHH421 pKa = 7.32LEE423 pKa = 4.08TEE425 pKa = 4.16DD426 pKa = 2.94QKK428 pKa = 11.97AAFRR432 pKa = 11.84DD433 pKa = 4.34FNLTGISQKK442 pKa = 10.53VADD445 pKa = 4.18VQSKK449 pKa = 8.24PVCDD453 pKa = 4.33AVLKK457 pKa = 8.53MLPAPHH463 pKa = 6.83IEE465 pKa = 4.18AKK467 pKa = 10.31ASLMKK472 pKa = 10.64GMSALDD478 pKa = 3.57VEE480 pKa = 5.13RR481 pKa = 11.84VLSASPEE488 pKa = 4.17DD489 pKa = 3.82EE490 pKa = 4.12VKK492 pKa = 10.69LLKK495 pKa = 10.37EE496 pKa = 4.04RR497 pKa = 11.84LRR499 pKa = 11.84GVDD502 pKa = 4.81PPCDD506 pKa = 3.48WAIHH510 pKa = 5.52DD511 pKa = 4.64AKK513 pKa = 10.65EE514 pKa = 3.67QAAEE518 pKa = 4.06AKK520 pKa = 10.3KK521 pKa = 10.63KK522 pKa = 10.26FYY524 pKa = 10.34QAAALEE530 pKa = 4.58LNTRR534 pKa = 11.84WEE536 pKa = 4.21KK537 pKa = 11.12AHH539 pKa = 5.91ATMMDD544 pKa = 4.93RR545 pKa = 11.84ITMEE549 pKa = 4.22ANAEE553 pKa = 4.09TRR555 pKa = 11.84QRR557 pKa = 11.84LAQGIQTLRR566 pKa = 11.84AEE568 pKa = 4.64LLRR571 pKa = 11.84LTTAEE576 pKa = 4.13EE577 pKa = 4.3MSGSDD582 pKa = 5.26LMKK585 pKa = 10.37QLQITYY591 pKa = 9.97RR592 pKa = 11.84ADD594 pKa = 4.4LEE596 pKa = 4.46EE597 pKa = 4.08HH598 pKa = 6.38WKK600 pKa = 11.05AGMAVIQKK608 pKa = 9.92LRR610 pKa = 11.84EE611 pKa = 3.94LL612 pKa = 4.02
Molecular weight: 68.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMP7|A0A1L3KMP7_9MONO Putative glycoprotein OS=Beihai barnacle virus 8 OX=1922366 PE=4 SV=1
MM1 pKa = 8.23DD2 pKa = 6.51DD3 pKa = 4.21LPEE6 pKa = 4.34HH7 pKa = 6.34LKK9 pKa = 10.15EE10 pKa = 4.25LKK12 pKa = 10.55VLADD16 pKa = 3.3HH17 pKa = 6.32KK18 pKa = 9.64TYY20 pKa = 10.73LKK22 pKa = 10.39KK23 pKa = 10.42PPSTGEE29 pKa = 4.18GTSSRR34 pKa = 11.84PEE36 pKa = 3.79SSPEE40 pKa = 3.71EE41 pKa = 3.87MFSASEE47 pKa = 4.13LSPSGQATPPASLRR61 pKa = 11.84TPLSGSQEE69 pKa = 4.25TLKK72 pKa = 10.88SHH74 pKa = 5.99IPVRR78 pKa = 11.84AGTTPPSSGQSLPPRR93 pKa = 11.84KK94 pKa = 9.62LSLHH98 pKa = 5.42SAEE101 pKa = 4.56CQTVEE106 pKa = 4.4PAHH109 pKa = 6.17SAASALLAWSGNYY122 pKa = 9.12KK123 pKa = 10.57KK124 pKa = 10.03PDD126 pKa = 3.12LVAAYY131 pKa = 7.36EE132 pKa = 4.04AWALNSTAPWKK143 pKa = 10.39KK144 pKa = 9.71ISGMYY149 pKa = 10.21KK150 pKa = 9.91KK151 pKa = 10.38FFEE154 pKa = 4.75AFPSYY159 pKa = 10.41SIKK162 pKa = 10.64EE163 pKa = 4.13PLSLKK168 pKa = 9.77PPKK171 pKa = 10.66GSDD174 pKa = 3.13FSQEE178 pKa = 3.39VWNTILSFIWEE189 pKa = 4.39DD190 pKa = 3.69CLDD193 pKa = 3.85NLIFCPRR200 pKa = 11.84EE201 pKa = 4.12MKK203 pKa = 10.4LHH205 pKa = 6.83WYY207 pKa = 10.18ACIAPPAEE215 pKa = 4.39CNKK218 pKa = 10.73SKK220 pKa = 10.68DD221 pKa = 3.46AHH223 pKa = 6.14CRR225 pKa = 11.84LAARR229 pKa = 11.84HH230 pKa = 5.02LHH232 pKa = 4.53VLKK235 pKa = 10.54MSLYY239 pKa = 9.75PSDD242 pKa = 3.72ATEE245 pKa = 3.61SDD247 pKa = 3.84YY248 pKa = 11.66LDD250 pKa = 4.21RR251 pKa = 11.84SPMTQLGEE259 pKa = 4.34SVMLLSSAGGLCRR272 pKa = 11.84AAKK275 pKa = 9.84AVSEE279 pKa = 4.42STHH282 pKa = 5.24QQVAMCKK289 pKa = 10.11DD290 pKa = 3.15AAAHH294 pKa = 4.81QKK296 pKa = 10.4IMLDD300 pKa = 3.2AATDD304 pKa = 3.47KK305 pKa = 9.1MTEE308 pKa = 3.73TSLYY312 pKa = 9.61CSQIMDD318 pKa = 4.37HH319 pKa = 6.81KK320 pKa = 10.16AQHH323 pKa = 6.44LSRR326 pKa = 11.84YY327 pKa = 9.88LDD329 pKa = 3.38LTMKK333 pKa = 10.55KK334 pKa = 10.52DD335 pKa = 3.9DD336 pKa = 3.63VDD338 pKa = 3.49TGKK341 pKa = 10.78FMAGSASVVTAPSEE355 pKa = 4.22RR356 pKa = 11.84LSSTSRR362 pKa = 11.84LSRR365 pKa = 11.84TSSAKK370 pKa = 10.39SSVSKK375 pKa = 9.87ATSAKK380 pKa = 9.77RR381 pKa = 11.84AQTGRR386 pKa = 11.84ATFPQVQKK394 pKa = 11.19LSS396 pKa = 3.58
MM1 pKa = 8.23DD2 pKa = 6.51DD3 pKa = 4.21LPEE6 pKa = 4.34HH7 pKa = 6.34LKK9 pKa = 10.15EE10 pKa = 4.25LKK12 pKa = 10.55VLADD16 pKa = 3.3HH17 pKa = 6.32KK18 pKa = 9.64TYY20 pKa = 10.73LKK22 pKa = 10.39KK23 pKa = 10.42PPSTGEE29 pKa = 4.18GTSSRR34 pKa = 11.84PEE36 pKa = 3.79SSPEE40 pKa = 3.71EE41 pKa = 3.87MFSASEE47 pKa = 4.13LSPSGQATPPASLRR61 pKa = 11.84TPLSGSQEE69 pKa = 4.25TLKK72 pKa = 10.88SHH74 pKa = 5.99IPVRR78 pKa = 11.84AGTTPPSSGQSLPPRR93 pKa = 11.84KK94 pKa = 9.62LSLHH98 pKa = 5.42SAEE101 pKa = 4.56CQTVEE106 pKa = 4.4PAHH109 pKa = 6.17SAASALLAWSGNYY122 pKa = 9.12KK123 pKa = 10.57KK124 pKa = 10.03PDD126 pKa = 3.12LVAAYY131 pKa = 7.36EE132 pKa = 4.04AWALNSTAPWKK143 pKa = 10.39KK144 pKa = 9.71ISGMYY149 pKa = 10.21KK150 pKa = 9.91KK151 pKa = 10.38FFEE154 pKa = 4.75AFPSYY159 pKa = 10.41SIKK162 pKa = 10.64EE163 pKa = 4.13PLSLKK168 pKa = 9.77PPKK171 pKa = 10.66GSDD174 pKa = 3.13FSQEE178 pKa = 3.39VWNTILSFIWEE189 pKa = 4.39DD190 pKa = 3.69CLDD193 pKa = 3.85NLIFCPRR200 pKa = 11.84EE201 pKa = 4.12MKK203 pKa = 10.4LHH205 pKa = 6.83WYY207 pKa = 10.18ACIAPPAEE215 pKa = 4.39CNKK218 pKa = 10.73SKK220 pKa = 10.68DD221 pKa = 3.46AHH223 pKa = 6.14CRR225 pKa = 11.84LAARR229 pKa = 11.84HH230 pKa = 5.02LHH232 pKa = 4.53VLKK235 pKa = 10.54MSLYY239 pKa = 9.75PSDD242 pKa = 3.72ATEE245 pKa = 3.61SDD247 pKa = 3.84YY248 pKa = 11.66LDD250 pKa = 4.21RR251 pKa = 11.84SPMTQLGEE259 pKa = 4.34SVMLLSSAGGLCRR272 pKa = 11.84AAKK275 pKa = 9.84AVSEE279 pKa = 4.42STHH282 pKa = 5.24QQVAMCKK289 pKa = 10.11DD290 pKa = 3.15AAAHH294 pKa = 4.81QKK296 pKa = 10.4IMLDD300 pKa = 3.2AATDD304 pKa = 3.47KK305 pKa = 9.1MTEE308 pKa = 3.73TSLYY312 pKa = 9.61CSQIMDD318 pKa = 4.37HH319 pKa = 6.81KK320 pKa = 10.16AQHH323 pKa = 6.44LSRR326 pKa = 11.84YY327 pKa = 9.88LDD329 pKa = 3.38LTMKK333 pKa = 10.55KK334 pKa = 10.52DD335 pKa = 3.9DD336 pKa = 3.63VDD338 pKa = 3.49TGKK341 pKa = 10.78FMAGSASVVTAPSEE355 pKa = 4.22RR356 pKa = 11.84LSSTSRR362 pKa = 11.84LSRR365 pKa = 11.84TSSAKK370 pKa = 10.39SSVSKK375 pKa = 9.87ATSAKK380 pKa = 9.77RR381 pKa = 11.84AQTGRR386 pKa = 11.84ATFPQVQKK394 pKa = 11.19LSS396 pKa = 3.58
Molecular weight: 43.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3854 |
396 |
2268 |
963.5 |
106.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.626 ± 0.564 | 1.505 ± 0.27 |
4.826 ± 0.269 | 6.046 ± 0.387 |
2.984 ± 0.181 | 5.864 ± 0.464 |
3.14 ± 0.15 | 4.307 ± 0.543 |
4.281 ± 0.846 | 11.417 ± 0.791 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.361 ± 0.319 | 2.309 ± 0.271 |
5.942 ± 0.566 | 4.463 ± 0.479 |
6.668 ± 0.701 | 8.329 ± 1.349 |
5.864 ± 0.67 | 5.838 ± 0.49 |
1.842 ± 0.198 | 2.387 ± 0.074 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
