
Halobellus clavatus
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Halobellus
Average proteome isoelectric point is 4.93
Get precalculated fractions of proteins
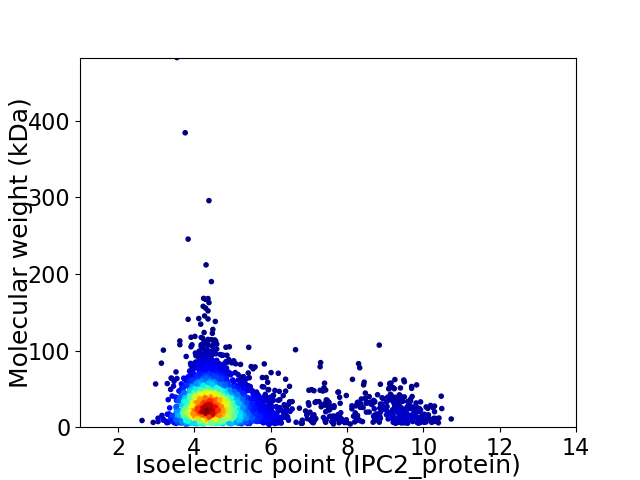
Virtual 2D-PAGE plot for 3679 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
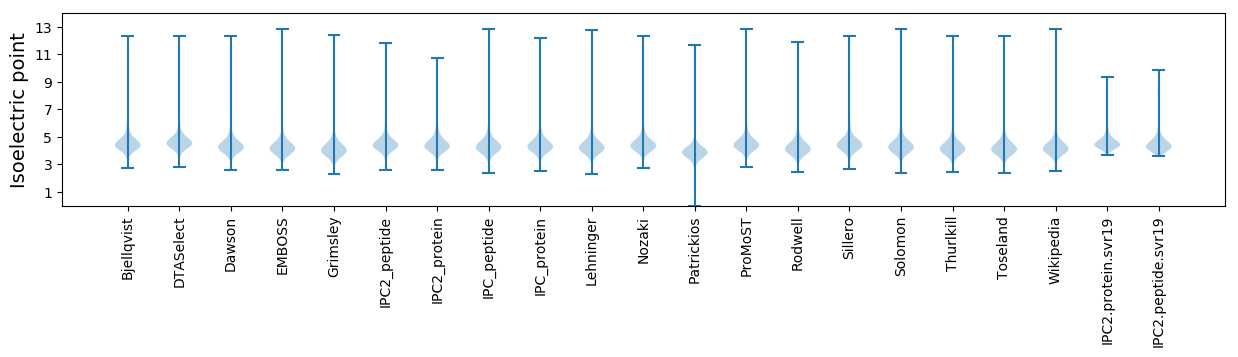
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3DKV8|A0A1H3DKV8_9EURY Uncharacterized protein OS=Halobellus clavatus OX=660517 GN=SAMN04487946_101600 PE=4 SV=1
MM1 pKa = 7.17NAEE4 pKa = 4.75DD5 pKa = 5.65FPTPDD10 pKa = 3.83ADD12 pKa = 3.99VEE14 pKa = 4.87SVTPEE19 pKa = 3.78ALKK22 pKa = 10.91DD23 pKa = 4.14RR24 pKa = 11.84IDD26 pKa = 3.66SGEE29 pKa = 4.38DD30 pKa = 3.07VTILDD35 pKa = 3.74TRR37 pKa = 11.84MQSDD41 pKa = 3.99YY42 pKa = 11.27EE43 pKa = 3.99EE44 pKa = 3.91WHH46 pKa = 6.65IEE48 pKa = 4.03GEE50 pKa = 4.47NVTSINVPYY59 pKa = 10.66FEE61 pKa = 5.8FLDD64 pKa = 3.8EE65 pKa = 6.63DD66 pKa = 3.49IDD68 pKa = 5.64ADD70 pKa = 4.0VLDD73 pKa = 5.41SIPADD78 pKa = 3.27EE79 pKa = 5.19HH80 pKa = 5.44VTVLCAKK87 pKa = 9.84GGASEE92 pKa = 4.41YY93 pKa = 10.77VAGTLVEE100 pKa = 3.96RR101 pKa = 11.84GYY103 pKa = 11.19DD104 pKa = 3.49VDD106 pKa = 3.9HH107 pKa = 7.49LEE109 pKa = 5.55DD110 pKa = 5.15GMNGWASIYY119 pKa = 9.98EE120 pKa = 4.11RR121 pKa = 11.84EE122 pKa = 4.26TVSRR126 pKa = 11.84YY127 pKa = 10.32DD128 pKa = 3.39GAGTLYY134 pKa = 10.35QYY136 pKa = 10.4QRR138 pKa = 11.84PSSGCLGYY146 pKa = 11.19LLVSDD151 pKa = 4.94GEE153 pKa = 4.22AAIVDD158 pKa = 4.1PLRR161 pKa = 11.84AFTDD165 pKa = 3.97RR166 pKa = 11.84YY167 pKa = 10.39LADD170 pKa = 4.15AADD173 pKa = 4.22LGADD177 pKa = 4.07LVYY180 pKa = 11.09ALDD183 pKa = 3.73THH185 pKa = 6.06VHH187 pKa = 6.6ADD189 pKa = 3.66HH190 pKa = 7.01ISGVRR195 pKa = 11.84DD196 pKa = 3.35LDD198 pKa = 4.0DD199 pKa = 4.43EE200 pKa = 4.52GVEE203 pKa = 4.4GVIPAASVDD212 pKa = 3.63RR213 pKa = 11.84GVTYY217 pKa = 10.96ADD219 pKa = 3.69EE220 pKa = 4.35LTNAEE225 pKa = 4.68DD226 pKa = 4.22GDD228 pKa = 4.27VFTVGDD234 pKa = 3.99AEE236 pKa = 4.41VEE238 pKa = 4.36TVYY241 pKa = 10.9TPGHH245 pKa = 4.66TTGMTSYY252 pKa = 11.22LLDD255 pKa = 4.93DD256 pKa = 4.4SLLATGDD263 pKa = 3.65GLFIEE268 pKa = 5.27SVARR272 pKa = 11.84PDD274 pKa = 4.69LEE276 pKa = 4.65EE277 pKa = 6.06GDD279 pKa = 4.23EE280 pKa = 4.34GAPDD284 pKa = 4.07AARR287 pKa = 11.84MLYY290 pKa = 10.33EE291 pKa = 4.13SLQEE295 pKa = 4.15RR296 pKa = 11.84VLTLPDD302 pKa = 3.46DD303 pKa = 4.06TLVGGAHH310 pKa = 6.6FSGAAVAAEE319 pKa = 4.55DD320 pKa = 3.79GTYY323 pKa = 9.56TATIGEE329 pKa = 4.83LKK331 pKa = 9.98TRR333 pKa = 11.84MDD335 pKa = 4.34ALSMDD340 pKa = 3.82EE341 pKa = 4.69EE342 pKa = 4.83DD343 pKa = 5.56FVDD346 pKa = 6.74LILADD351 pKa = 3.98MPPRR355 pKa = 11.84PANYY359 pKa = 9.92EE360 pKa = 4.11DD361 pKa = 5.2IIATNLGQHH370 pKa = 6.35AVDD373 pKa = 4.37DD374 pKa = 4.79DD375 pKa = 4.19EE376 pKa = 6.15AFSLEE381 pKa = 4.58LGPNNCAASQEE392 pKa = 4.43SLAGDD397 pKa = 3.77
MM1 pKa = 7.17NAEE4 pKa = 4.75DD5 pKa = 5.65FPTPDD10 pKa = 3.83ADD12 pKa = 3.99VEE14 pKa = 4.87SVTPEE19 pKa = 3.78ALKK22 pKa = 10.91DD23 pKa = 4.14RR24 pKa = 11.84IDD26 pKa = 3.66SGEE29 pKa = 4.38DD30 pKa = 3.07VTILDD35 pKa = 3.74TRR37 pKa = 11.84MQSDD41 pKa = 3.99YY42 pKa = 11.27EE43 pKa = 3.99EE44 pKa = 3.91WHH46 pKa = 6.65IEE48 pKa = 4.03GEE50 pKa = 4.47NVTSINVPYY59 pKa = 10.66FEE61 pKa = 5.8FLDD64 pKa = 3.8EE65 pKa = 6.63DD66 pKa = 3.49IDD68 pKa = 5.64ADD70 pKa = 4.0VLDD73 pKa = 5.41SIPADD78 pKa = 3.27EE79 pKa = 5.19HH80 pKa = 5.44VTVLCAKK87 pKa = 9.84GGASEE92 pKa = 4.41YY93 pKa = 10.77VAGTLVEE100 pKa = 3.96RR101 pKa = 11.84GYY103 pKa = 11.19DD104 pKa = 3.49VDD106 pKa = 3.9HH107 pKa = 7.49LEE109 pKa = 5.55DD110 pKa = 5.15GMNGWASIYY119 pKa = 9.98EE120 pKa = 4.11RR121 pKa = 11.84EE122 pKa = 4.26TVSRR126 pKa = 11.84YY127 pKa = 10.32DD128 pKa = 3.39GAGTLYY134 pKa = 10.35QYY136 pKa = 10.4QRR138 pKa = 11.84PSSGCLGYY146 pKa = 11.19LLVSDD151 pKa = 4.94GEE153 pKa = 4.22AAIVDD158 pKa = 4.1PLRR161 pKa = 11.84AFTDD165 pKa = 3.97RR166 pKa = 11.84YY167 pKa = 10.39LADD170 pKa = 4.15AADD173 pKa = 4.22LGADD177 pKa = 4.07LVYY180 pKa = 11.09ALDD183 pKa = 3.73THH185 pKa = 6.06VHH187 pKa = 6.6ADD189 pKa = 3.66HH190 pKa = 7.01ISGVRR195 pKa = 11.84DD196 pKa = 3.35LDD198 pKa = 4.0DD199 pKa = 4.43EE200 pKa = 4.52GVEE203 pKa = 4.4GVIPAASVDD212 pKa = 3.63RR213 pKa = 11.84GVTYY217 pKa = 10.96ADD219 pKa = 3.69EE220 pKa = 4.35LTNAEE225 pKa = 4.68DD226 pKa = 4.22GDD228 pKa = 4.27VFTVGDD234 pKa = 3.99AEE236 pKa = 4.41VEE238 pKa = 4.36TVYY241 pKa = 10.9TPGHH245 pKa = 4.66TTGMTSYY252 pKa = 11.22LLDD255 pKa = 4.93DD256 pKa = 4.4SLLATGDD263 pKa = 3.65GLFIEE268 pKa = 5.27SVARR272 pKa = 11.84PDD274 pKa = 4.69LEE276 pKa = 4.65EE277 pKa = 6.06GDD279 pKa = 4.23EE280 pKa = 4.34GAPDD284 pKa = 4.07AARR287 pKa = 11.84MLYY290 pKa = 10.33EE291 pKa = 4.13SLQEE295 pKa = 4.15RR296 pKa = 11.84VLTLPDD302 pKa = 3.46DD303 pKa = 4.06TLVGGAHH310 pKa = 6.6FSGAAVAAEE319 pKa = 4.55DD320 pKa = 3.79GTYY323 pKa = 9.56TATIGEE329 pKa = 4.83LKK331 pKa = 9.98TRR333 pKa = 11.84MDD335 pKa = 4.34ALSMDD340 pKa = 3.82EE341 pKa = 4.69EE342 pKa = 4.83DD343 pKa = 5.56FVDD346 pKa = 6.74LILADD351 pKa = 3.98MPPRR355 pKa = 11.84PANYY359 pKa = 9.92EE360 pKa = 4.11DD361 pKa = 5.2IIATNLGQHH370 pKa = 6.35AVDD373 pKa = 4.37DD374 pKa = 4.79DD375 pKa = 4.19EE376 pKa = 6.15AFSLEE381 pKa = 4.58LGPNNCAASQEE392 pKa = 4.43SLAGDD397 pKa = 3.77
Molecular weight: 42.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3IDS0|A0A1H3IDS0_9EURY O-succinylbenzoic acid--CoA ligase OS=Halobellus clavatus OX=660517 GN=SAMN04487946_109124 PE=4 SV=1
MM1 pKa = 7.43SATPVVVALSLVSGAFWGVGPIFSKK26 pKa = 11.01LGMEE30 pKa = 4.4RR31 pKa = 11.84GGRR34 pKa = 11.84PHH36 pKa = 7.54RR37 pKa = 11.84ATLIVLVVGAAVFWAALLVRR57 pKa = 11.84GAGQASVARR66 pKa = 11.84LPAVDD71 pKa = 3.05ITVFAVSGVIGTSLAWLSWFRR92 pKa = 11.84GIRR95 pKa = 11.84LVGASVGNVLFYY107 pKa = 9.14TQPLFATLLAAAVLGEE123 pKa = 4.34RR124 pKa = 11.84LTSVTGVGVVLIVLGVGLLSLSGGSNLEE152 pKa = 3.81FRR154 pKa = 11.84LSKK157 pKa = 10.99ALLFPLAAAVFAAISNVVNRR177 pKa = 11.84FGFQTSTATPLEE189 pKa = 4.11AAAINLTSALPFIVGHH205 pKa = 4.85TLGYY209 pKa = 9.91HH210 pKa = 6.06RR211 pKa = 11.84NALGTPDD218 pKa = 4.38RR219 pKa = 11.84SDD221 pKa = 3.47AYY223 pKa = 9.62FVCSGLANAVAVFTMFTALANGPVVIVAPLVSTSPLFTTALAAVFLTDD271 pKa = 3.9VEE273 pKa = 4.88TITARR278 pKa = 11.84TVASAVLTVVGAGITAVV295 pKa = 3.22
MM1 pKa = 7.43SATPVVVALSLVSGAFWGVGPIFSKK26 pKa = 11.01LGMEE30 pKa = 4.4RR31 pKa = 11.84GGRR34 pKa = 11.84PHH36 pKa = 7.54RR37 pKa = 11.84ATLIVLVVGAAVFWAALLVRR57 pKa = 11.84GAGQASVARR66 pKa = 11.84LPAVDD71 pKa = 3.05ITVFAVSGVIGTSLAWLSWFRR92 pKa = 11.84GIRR95 pKa = 11.84LVGASVGNVLFYY107 pKa = 9.14TQPLFATLLAAAVLGEE123 pKa = 4.34RR124 pKa = 11.84LTSVTGVGVVLIVLGVGLLSLSGGSNLEE152 pKa = 3.81FRR154 pKa = 11.84LSKK157 pKa = 10.99ALLFPLAAAVFAAISNVVNRR177 pKa = 11.84FGFQTSTATPLEE189 pKa = 4.11AAAINLTSALPFIVGHH205 pKa = 4.85TLGYY209 pKa = 9.91HH210 pKa = 6.06RR211 pKa = 11.84NALGTPDD218 pKa = 4.38RR219 pKa = 11.84SDD221 pKa = 3.47AYY223 pKa = 9.62FVCSGLANAVAVFTMFTALANGPVVIVAPLVSTSPLFTTALAAVFLTDD271 pKa = 3.9VEE273 pKa = 4.88TITARR278 pKa = 11.84TVASAVLTVVGAGITAVV295 pKa = 3.22
Molecular weight: 30.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1088495 |
39 |
4622 |
295.9 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.89 ± 0.061 | 0.656 ± 0.013 |
8.367 ± 0.048 | 8.387 ± 0.059 |
3.286 ± 0.029 | 8.36 ± 0.043 |
1.953 ± 0.023 | 4.256 ± 0.032 |
1.845 ± 0.034 | 8.873 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.018 | 2.412 ± 0.031 |
4.708 ± 0.03 | 2.697 ± 0.023 |
6.634 ± 0.042 | 5.894 ± 0.037 |
6.548 ± 0.058 | 8.806 ± 0.046 |
1.122 ± 0.018 | 2.694 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
