
Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / CCUG 37298 / CIP 103539 / LMG 7603 / PAl5)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconacetobacter; Gluconacetobacter diazotrophicus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
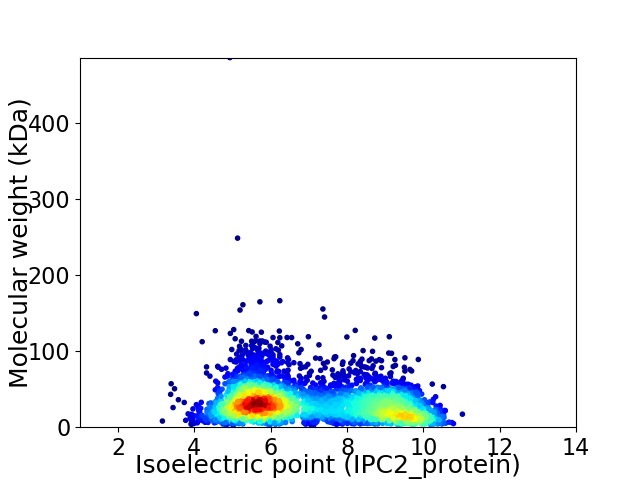
Virtual 2D-PAGE plot for 3783 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
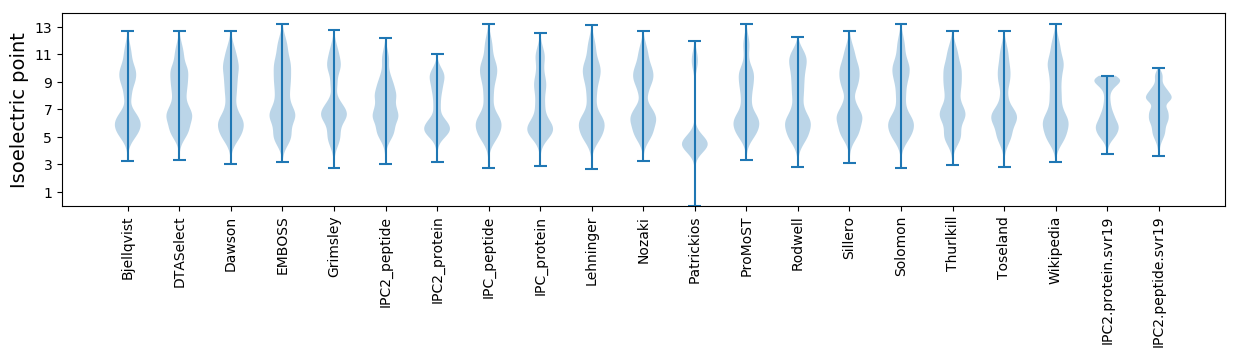
* You can choose from 21 different methods for calculating isoelectric point
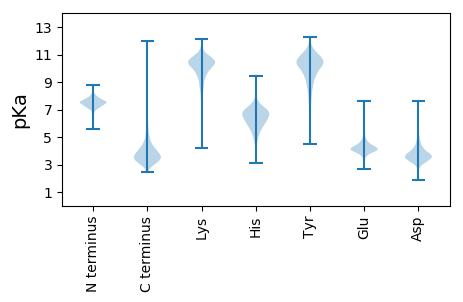
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9H7G9|A9H7G9_GLUDA Uncharacterized protein OS=Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / CCUG 37298 / CIP 103539 / LMG 7603 / PAl5) OX=272568 GN=GDI3695 PE=4 SV=1
MM1 pKa = 7.37SGSAARR7 pKa = 11.84FVWPNSFSVDD17 pKa = 3.11AAGVPRR23 pKa = 11.84AGARR27 pKa = 11.84LFFYY31 pKa = 9.65QTGTTTPQATYY42 pKa = 10.78ADD44 pKa = 4.06AGLTVPNANPVIADD58 pKa = 3.53EE59 pKa = 4.43NGQFGSIFLLGAPSYY74 pKa = 11.13AVALEE79 pKa = 4.5DD80 pKa = 5.08GEE82 pKa = 5.02GNPVWTMDD90 pKa = 3.7PVGAAGGGVGAVPVGTVADD109 pKa = 4.08YY110 pKa = 11.58AGATAPAGG118 pKa = 3.56
MM1 pKa = 7.37SGSAARR7 pKa = 11.84FVWPNSFSVDD17 pKa = 3.11AAGVPRR23 pKa = 11.84AGARR27 pKa = 11.84LFFYY31 pKa = 9.65QTGTTTPQATYY42 pKa = 10.78ADD44 pKa = 4.06AGLTVPNANPVIADD58 pKa = 3.53EE59 pKa = 4.43NGQFGSIFLLGAPSYY74 pKa = 11.13AVALEE79 pKa = 4.5DD80 pKa = 5.08GEE82 pKa = 5.02GNPVWTMDD90 pKa = 3.7PVGAAGGGVGAVPVGTVADD109 pKa = 4.08YY110 pKa = 11.58AGATAPAGG118 pKa = 3.56
Molecular weight: 11.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9HQC7|A9HQC7_GLUDA Putative alkyl hydroperoxide reductase OS=Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / CCUG 37298 / CIP 103539 / LMG 7603 / PAl5) OX=272568 GN=ahpC PE=4 SV=1
MM1 pKa = 6.55TTPPRR6 pKa = 11.84RR7 pKa = 11.84PCSSPRR13 pKa = 11.84SWQRR17 pKa = 11.84HH18 pKa = 4.06RR19 pKa = 11.84QPGPFRR25 pKa = 11.84RR26 pKa = 11.84CRR28 pKa = 11.84PPHH31 pKa = 5.61FRR33 pKa = 11.84HH34 pKa = 6.44RR35 pKa = 11.84PLPPLQRR42 pKa = 11.84CKK44 pKa = 10.53PSWQHH49 pKa = 5.71PRR51 pKa = 11.84RR52 pKa = 11.84LPLRR56 pKa = 11.84VRR58 pKa = 11.84RR59 pKa = 11.84LPSRR63 pKa = 11.84QVRR66 pKa = 11.84QPRR69 pKa = 11.84PTPGQRR75 pKa = 11.84GQPRR79 pKa = 11.84QRR81 pKa = 11.84LPSHH85 pKa = 6.86RR86 pKa = 11.84PPFQSRR92 pKa = 11.84PPPRR96 pKa = 11.84RR97 pKa = 11.84QAAAHH102 pKa = 6.2RR103 pKa = 11.84KK104 pKa = 6.63SARR107 pKa = 11.84WRR109 pKa = 11.84HH110 pKa = 5.67CRR112 pKa = 11.84HH113 pKa = 4.84TWRR116 pKa = 11.84AQSSRR121 pKa = 11.84GRR123 pKa = 11.84RR124 pKa = 11.84APPRR128 pKa = 11.84APGRR132 pKa = 11.84SLAGYY137 pKa = 9.27RR138 pKa = 11.84RR139 pKa = 11.84KK140 pKa = 7.8TWW142 pKa = 2.86
MM1 pKa = 6.55TTPPRR6 pKa = 11.84RR7 pKa = 11.84PCSSPRR13 pKa = 11.84SWQRR17 pKa = 11.84HH18 pKa = 4.06RR19 pKa = 11.84QPGPFRR25 pKa = 11.84RR26 pKa = 11.84CRR28 pKa = 11.84PPHH31 pKa = 5.61FRR33 pKa = 11.84HH34 pKa = 6.44RR35 pKa = 11.84PLPPLQRR42 pKa = 11.84CKK44 pKa = 10.53PSWQHH49 pKa = 5.71PRR51 pKa = 11.84RR52 pKa = 11.84LPLRR56 pKa = 11.84VRR58 pKa = 11.84RR59 pKa = 11.84LPSRR63 pKa = 11.84QVRR66 pKa = 11.84QPRR69 pKa = 11.84PTPGQRR75 pKa = 11.84GQPRR79 pKa = 11.84QRR81 pKa = 11.84LPSHH85 pKa = 6.86RR86 pKa = 11.84PPFQSRR92 pKa = 11.84PPPRR96 pKa = 11.84RR97 pKa = 11.84QAAAHH102 pKa = 6.2RR103 pKa = 11.84KK104 pKa = 6.63SARR107 pKa = 11.84WRR109 pKa = 11.84HH110 pKa = 5.67CRR112 pKa = 11.84HH113 pKa = 4.84TWRR116 pKa = 11.84AQSSRR121 pKa = 11.84GRR123 pKa = 11.84RR124 pKa = 11.84APPRR128 pKa = 11.84APGRR132 pKa = 11.84SLAGYY137 pKa = 9.27RR138 pKa = 11.84RR139 pKa = 11.84KK140 pKa = 7.8TWW142 pKa = 2.86
Molecular weight: 17.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1180201 |
19 |
4453 |
312.0 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.989 ± 0.069 | 0.999 ± 0.013 |
5.879 ± 0.035 | 4.55 ± 0.039 |
3.332 ± 0.026 | 8.963 ± 0.042 |
2.4 ± 0.019 | 4.732 ± 0.029 |
2.108 ± 0.032 | 10.191 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.582 ± 0.018 | 2.255 ± 0.025 |
5.902 ± 0.037 | 3.181 ± 0.028 |
8.256 ± 0.046 | 5.181 ± 0.034 |
5.564 ± 0.033 | 7.424 ± 0.034 |
1.413 ± 0.017 | 2.098 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
