
Wenling thamnaconus septentrionalis filovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Thamnovirus; Huangjiao thamnovirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
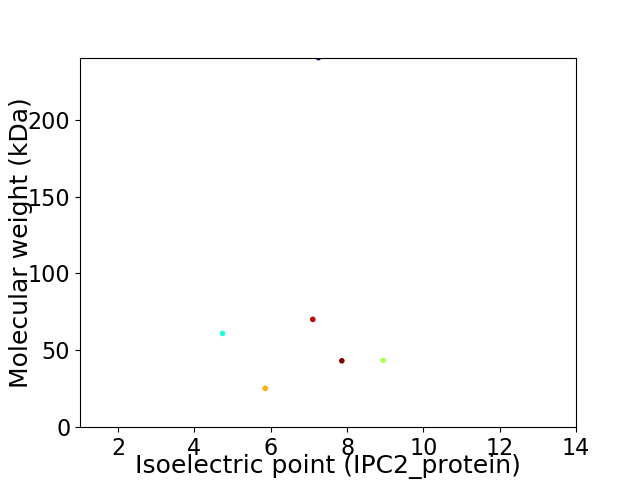
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GMP6|A0A2P1GMP6_9MONO Uncharacterized protein OS=Wenling thamnaconus septentrionalis filovirus OX=2116488 PE=4 SV=1
MM1 pKa = 6.84STSEE5 pKa = 4.4KK6 pKa = 10.64EE7 pKa = 3.71MANRR11 pKa = 11.84IMKK14 pKa = 10.59DD15 pKa = 2.43SDD17 pKa = 3.63LQVIGLLLDD26 pKa = 4.03GLDD29 pKa = 3.94DD30 pKa = 3.57QGLDD34 pKa = 3.51VRR36 pKa = 11.84ALPDD40 pKa = 3.3PTGNRR45 pKa = 11.84DD46 pKa = 3.47YY47 pKa = 11.41TDD49 pKa = 2.75VSTFKK54 pKa = 11.04DD55 pKa = 4.4LFPMMSTSEE64 pKa = 4.57LIGSLSASGYY74 pKa = 10.14SDD76 pKa = 3.4QLEE79 pKa = 4.12KK80 pKa = 10.91SIEE83 pKa = 4.09STGRR87 pKa = 11.84KK88 pKa = 9.04LADD91 pKa = 3.79LARR94 pKa = 11.84SAPADD99 pKa = 3.81DD100 pKa = 4.54NAVQFMKK107 pKa = 10.34SYY109 pKa = 11.48LLMCEE114 pKa = 3.99KK115 pKa = 9.96TGQEE119 pKa = 4.26HH120 pKa = 6.6SVSLPKK126 pKa = 10.45EE127 pKa = 3.78EE128 pKa = 4.54RR129 pKa = 11.84KK130 pKa = 10.4GEE132 pKa = 4.23KK133 pKa = 8.83KK134 pKa = 9.12TGEE137 pKa = 4.05HH138 pKa = 6.05MKK140 pKa = 10.28PEE142 pKa = 4.31QSQGPQPTDD151 pKa = 3.23KK152 pKa = 11.25NKK154 pKa = 8.95TAAKK158 pKa = 9.79AGPSEE163 pKa = 4.04TGKK166 pKa = 10.85ANEE169 pKa = 4.2EE170 pKa = 4.04EE171 pKa = 4.66VTNKK175 pKa = 9.21TPAPRR180 pKa = 11.84RR181 pKa = 11.84NTPKK185 pKa = 10.4ALSGAEE191 pKa = 4.31TNDD194 pKa = 3.82SPLSTQLTQTPEE206 pKa = 3.69EE207 pKa = 4.32AGDD210 pKa = 3.76MFSGKK215 pKa = 10.45AEE217 pKa = 4.6GIVSGEE223 pKa = 3.63LRR225 pKa = 11.84TGKK228 pKa = 10.22KK229 pKa = 10.24LPTIVEE235 pKa = 4.26GCDD238 pKa = 2.92ASGRR242 pKa = 11.84GDD244 pKa = 3.32VTHH247 pKa = 7.68LYY249 pKa = 10.08RR250 pKa = 11.84SHH252 pKa = 6.66EE253 pKa = 4.11LVEE256 pKa = 4.91RR257 pKa = 11.84IQKK260 pKa = 10.12GEE262 pKa = 3.75KK263 pKa = 9.57TMGIACLKK271 pKa = 9.27ALVMGEE277 pKa = 4.52DD278 pKa = 3.67NFSGVPFDD286 pKa = 6.75DD287 pKa = 4.45ILDD290 pKa = 4.29CFGEE294 pKa = 4.46TCDD297 pKa = 6.07DD298 pKa = 4.52PDD300 pKa = 3.93TPGLRR305 pKa = 11.84GLMEE309 pKa = 4.93GSNGPASGLPCTDD322 pKa = 4.96DD323 pKa = 3.39YY324 pKa = 11.63CGPSTVLFDD333 pKa = 3.72RR334 pKa = 11.84WAKK337 pKa = 10.5AGSLGCVNYY346 pKa = 9.8MIRR349 pKa = 11.84RR350 pKa = 11.84CMGDD354 pKa = 3.1LYY356 pKa = 11.28DD357 pKa = 4.28GLRR360 pKa = 11.84AAACDD365 pKa = 3.59TPAYY369 pKa = 7.57YY370 pKa = 10.41HH371 pKa = 7.26EE372 pKa = 4.81MAQQQKK378 pKa = 10.6DD379 pKa = 3.66CTDD382 pKa = 3.22QVVKK386 pKa = 10.64VVSGMEE392 pKa = 3.98EE393 pKa = 3.61MGEE396 pKa = 3.86AMQEE400 pKa = 3.74LRR402 pKa = 11.84RR403 pKa = 11.84EE404 pKa = 4.02NTEE407 pKa = 3.55MKK409 pKa = 10.45RR410 pKa = 11.84RR411 pKa = 11.84IADD414 pKa = 3.11MAAAMEE420 pKa = 4.41HH421 pKa = 6.15LSGVVSSLSSKK432 pKa = 11.01LEE434 pKa = 3.88TAVVVSSGYY443 pKa = 9.65PSSKK447 pKa = 8.62EE448 pKa = 3.49TAAGIKK454 pKa = 10.17GSTYY458 pKa = 11.05NEE460 pKa = 4.31DD461 pKa = 3.22QQTWKK466 pKa = 9.32TGVSSWQKK474 pKa = 10.26RR475 pKa = 11.84EE476 pKa = 3.69TSIWTNPSEE485 pKa = 5.41RR486 pKa = 11.84SPQEE490 pKa = 3.89LNTHH494 pKa = 6.09GGIGEE499 pKa = 4.56DD500 pKa = 4.23DD501 pKa = 3.77SEE503 pKa = 5.59AFLKK507 pKa = 10.46QLEE510 pKa = 4.43EE511 pKa = 3.63QWAAEE516 pKa = 4.1SSDD519 pKa = 3.48PLSFGTSGRR528 pKa = 11.84GINDD532 pKa = 2.88QDD534 pKa = 3.94FVRR537 pKa = 11.84EE538 pKa = 4.23TLTGTPYY545 pKa = 10.75EE546 pKa = 4.29GDD548 pKa = 3.56YY549 pKa = 10.8NASRR553 pKa = 11.84EE554 pKa = 4.44SKK556 pKa = 10.2KK557 pKa = 10.87GPRR560 pKa = 11.84FF561 pKa = 3.43
MM1 pKa = 6.84STSEE5 pKa = 4.4KK6 pKa = 10.64EE7 pKa = 3.71MANRR11 pKa = 11.84IMKK14 pKa = 10.59DD15 pKa = 2.43SDD17 pKa = 3.63LQVIGLLLDD26 pKa = 4.03GLDD29 pKa = 3.94DD30 pKa = 3.57QGLDD34 pKa = 3.51VRR36 pKa = 11.84ALPDD40 pKa = 3.3PTGNRR45 pKa = 11.84DD46 pKa = 3.47YY47 pKa = 11.41TDD49 pKa = 2.75VSTFKK54 pKa = 11.04DD55 pKa = 4.4LFPMMSTSEE64 pKa = 4.57LIGSLSASGYY74 pKa = 10.14SDD76 pKa = 3.4QLEE79 pKa = 4.12KK80 pKa = 10.91SIEE83 pKa = 4.09STGRR87 pKa = 11.84KK88 pKa = 9.04LADD91 pKa = 3.79LARR94 pKa = 11.84SAPADD99 pKa = 3.81DD100 pKa = 4.54NAVQFMKK107 pKa = 10.34SYY109 pKa = 11.48LLMCEE114 pKa = 3.99KK115 pKa = 9.96TGQEE119 pKa = 4.26HH120 pKa = 6.6SVSLPKK126 pKa = 10.45EE127 pKa = 3.78EE128 pKa = 4.54RR129 pKa = 11.84KK130 pKa = 10.4GEE132 pKa = 4.23KK133 pKa = 8.83KK134 pKa = 9.12TGEE137 pKa = 4.05HH138 pKa = 6.05MKK140 pKa = 10.28PEE142 pKa = 4.31QSQGPQPTDD151 pKa = 3.23KK152 pKa = 11.25NKK154 pKa = 8.95TAAKK158 pKa = 9.79AGPSEE163 pKa = 4.04TGKK166 pKa = 10.85ANEE169 pKa = 4.2EE170 pKa = 4.04EE171 pKa = 4.66VTNKK175 pKa = 9.21TPAPRR180 pKa = 11.84RR181 pKa = 11.84NTPKK185 pKa = 10.4ALSGAEE191 pKa = 4.31TNDD194 pKa = 3.82SPLSTQLTQTPEE206 pKa = 3.69EE207 pKa = 4.32AGDD210 pKa = 3.76MFSGKK215 pKa = 10.45AEE217 pKa = 4.6GIVSGEE223 pKa = 3.63LRR225 pKa = 11.84TGKK228 pKa = 10.22KK229 pKa = 10.24LPTIVEE235 pKa = 4.26GCDD238 pKa = 2.92ASGRR242 pKa = 11.84GDD244 pKa = 3.32VTHH247 pKa = 7.68LYY249 pKa = 10.08RR250 pKa = 11.84SHH252 pKa = 6.66EE253 pKa = 4.11LVEE256 pKa = 4.91RR257 pKa = 11.84IQKK260 pKa = 10.12GEE262 pKa = 3.75KK263 pKa = 9.57TMGIACLKK271 pKa = 9.27ALVMGEE277 pKa = 4.52DD278 pKa = 3.67NFSGVPFDD286 pKa = 6.75DD287 pKa = 4.45ILDD290 pKa = 4.29CFGEE294 pKa = 4.46TCDD297 pKa = 6.07DD298 pKa = 4.52PDD300 pKa = 3.93TPGLRR305 pKa = 11.84GLMEE309 pKa = 4.93GSNGPASGLPCTDD322 pKa = 4.96DD323 pKa = 3.39YY324 pKa = 11.63CGPSTVLFDD333 pKa = 3.72RR334 pKa = 11.84WAKK337 pKa = 10.5AGSLGCVNYY346 pKa = 9.8MIRR349 pKa = 11.84RR350 pKa = 11.84CMGDD354 pKa = 3.1LYY356 pKa = 11.28DD357 pKa = 4.28GLRR360 pKa = 11.84AAACDD365 pKa = 3.59TPAYY369 pKa = 7.57YY370 pKa = 10.41HH371 pKa = 7.26EE372 pKa = 4.81MAQQQKK378 pKa = 10.6DD379 pKa = 3.66CTDD382 pKa = 3.22QVVKK386 pKa = 10.64VVSGMEE392 pKa = 3.98EE393 pKa = 3.61MGEE396 pKa = 3.86AMQEE400 pKa = 3.74LRR402 pKa = 11.84RR403 pKa = 11.84EE404 pKa = 4.02NTEE407 pKa = 3.55MKK409 pKa = 10.45RR410 pKa = 11.84RR411 pKa = 11.84IADD414 pKa = 3.11MAAAMEE420 pKa = 4.41HH421 pKa = 6.15LSGVVSSLSSKK432 pKa = 11.01LEE434 pKa = 3.88TAVVVSSGYY443 pKa = 9.65PSSKK447 pKa = 8.62EE448 pKa = 3.49TAAGIKK454 pKa = 10.17GSTYY458 pKa = 11.05NEE460 pKa = 4.31DD461 pKa = 3.22QQTWKK466 pKa = 9.32TGVSSWQKK474 pKa = 10.26RR475 pKa = 11.84EE476 pKa = 3.69TSIWTNPSEE485 pKa = 5.41RR486 pKa = 11.84SPQEE490 pKa = 3.89LNTHH494 pKa = 6.09GGIGEE499 pKa = 4.56DD500 pKa = 4.23DD501 pKa = 3.77SEE503 pKa = 5.59AFLKK507 pKa = 10.46QLEE510 pKa = 4.43EE511 pKa = 3.63QWAAEE516 pKa = 4.1SSDD519 pKa = 3.48PLSFGTSGRR528 pKa = 11.84GINDD532 pKa = 2.88QDD534 pKa = 3.94FVRR537 pKa = 11.84EE538 pKa = 4.23TLTGTPYY545 pKa = 10.75EE546 pKa = 4.29GDD548 pKa = 3.56YY549 pKa = 10.8NASRR553 pKa = 11.84EE554 pKa = 4.44SKK556 pKa = 10.2KK557 pKa = 10.87GPRR560 pKa = 11.84FF561 pKa = 3.43
Molecular weight: 60.97 kDa
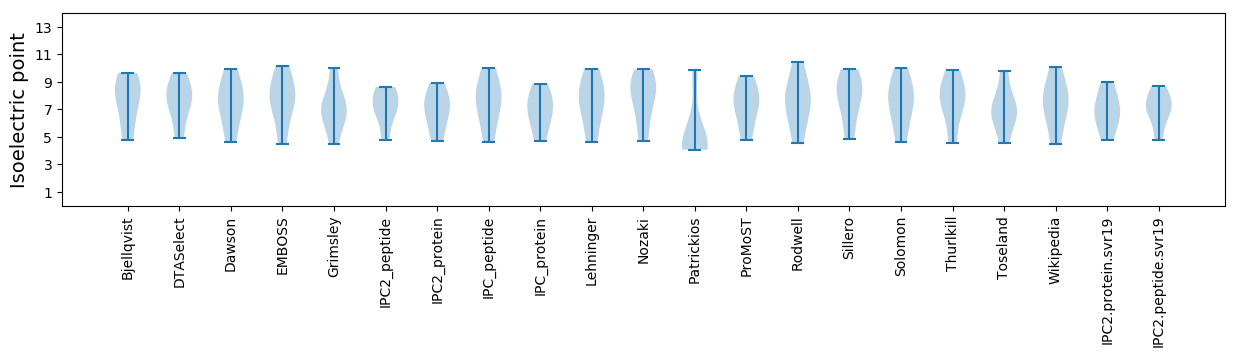
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GMM5|A0A2P1GMM5_9MONO Nucleoprotein OS=Wenling thamnaconus septentrionalis filovirus OX=2116488 PE=4 SV=1
MM1 pKa = 6.89ITNLITTSLRR11 pKa = 11.84KK12 pKa = 9.56KK13 pKa = 9.78KK14 pKa = 10.29RR15 pKa = 11.84KK16 pKa = 8.58EE17 pKa = 3.61KK18 pKa = 10.75RR19 pKa = 11.84KK20 pKa = 7.92TKK22 pKa = 10.29PIGGNMASKK31 pKa = 9.89PVRR34 pKa = 11.84GGGSGGGSGHH44 pKa = 7.01RR45 pKa = 11.84PFHH48 pKa = 6.53PNQLPQYY55 pKa = 8.37KK56 pKa = 9.65ARR58 pKa = 11.84SRR60 pKa = 11.84ICYY63 pKa = 9.31GFLSGTCKK71 pKa = 10.3RR72 pKa = 11.84GISCRR77 pKa = 11.84YY78 pKa = 7.81QHH80 pKa = 6.85DD81 pKa = 4.36LKK83 pKa = 11.26CGGEE87 pKa = 4.04RR88 pKa = 11.84EE89 pKa = 4.03QLLFSRR95 pKa = 11.84MKK97 pKa = 10.51VDD99 pKa = 4.16QLLLANSKK107 pKa = 9.72GRR109 pKa = 11.84APQMVKK115 pKa = 10.93DD116 pKa = 4.0NLLSGVLNEE125 pKa = 4.08IKK127 pKa = 10.52KK128 pKa = 10.13RR129 pKa = 11.84VPKK132 pKa = 10.77SEE134 pKa = 4.04DD135 pKa = 3.07RR136 pKa = 11.84CTLGDD141 pKa = 3.9SDD143 pKa = 5.74LKK145 pKa = 10.56TMLNGFEE152 pKa = 4.43KK153 pKa = 11.1VNAGLNNPQRR163 pKa = 11.84LEE165 pKa = 3.82SWDD168 pKa = 3.48PSGTVDD174 pKa = 5.27LGRR177 pKa = 11.84LSLLGNPQRR186 pKa = 11.84LGWRR190 pKa = 11.84LGHH193 pKa = 7.2LKK195 pKa = 10.67ALTCAQLVILLDD207 pKa = 3.9FLVKK211 pKa = 10.59ALTDD215 pKa = 3.57MEE217 pKa = 4.6EE218 pKa = 4.18EE219 pKa = 4.42GSEE222 pKa = 4.64GEE224 pKa = 4.13GSLWGARR231 pKa = 11.84HH232 pKa = 5.1EE233 pKa = 4.37HH234 pKa = 6.25SSRR237 pKa = 11.84GCRR240 pKa = 11.84YY241 pKa = 9.11YY242 pKa = 11.01KK243 pKa = 10.51SGMCFKK249 pKa = 10.21MGRR252 pKa = 11.84CLYY255 pKa = 10.33SHH257 pKa = 6.92NPYY260 pKa = 10.8KK261 pKa = 10.83LGASDD266 pKa = 5.52LIRR269 pKa = 11.84GIAIGLQSLDD279 pKa = 4.37LGSQPLCEE287 pKa = 4.4DD288 pKa = 3.09SGMPSLPQDD297 pKa = 3.04RR298 pKa = 11.84KK299 pKa = 10.28ARR301 pKa = 11.84KK302 pKa = 9.69DD303 pKa = 3.15MARR306 pKa = 11.84ALISEE311 pKa = 4.41SAINPRR317 pKa = 11.84LPKK320 pKa = 10.44EE321 pKa = 4.26SLLTMAQLIWEE332 pKa = 4.36WDD334 pKa = 3.56YY335 pKa = 11.88VPPSEE340 pKa = 5.53SGPTPPSSGDD350 pKa = 3.23VPGEE354 pKa = 3.93VRR356 pKa = 11.84DD357 pKa = 3.77MLLTLKK363 pKa = 10.73ALEE366 pKa = 4.52GPKK369 pKa = 10.4GVAAALTRR377 pKa = 11.84YY378 pKa = 8.72SQEE381 pKa = 3.32HH382 pKa = 6.69RR383 pKa = 11.84GRR385 pKa = 11.84LLISMEE391 pKa = 3.99KK392 pKa = 10.6KK393 pKa = 8.95MM394 pKa = 4.99
MM1 pKa = 6.89ITNLITTSLRR11 pKa = 11.84KK12 pKa = 9.56KK13 pKa = 9.78KK14 pKa = 10.29RR15 pKa = 11.84KK16 pKa = 8.58EE17 pKa = 3.61KK18 pKa = 10.75RR19 pKa = 11.84KK20 pKa = 7.92TKK22 pKa = 10.29PIGGNMASKK31 pKa = 9.89PVRR34 pKa = 11.84GGGSGGGSGHH44 pKa = 7.01RR45 pKa = 11.84PFHH48 pKa = 6.53PNQLPQYY55 pKa = 8.37KK56 pKa = 9.65ARR58 pKa = 11.84SRR60 pKa = 11.84ICYY63 pKa = 9.31GFLSGTCKK71 pKa = 10.3RR72 pKa = 11.84GISCRR77 pKa = 11.84YY78 pKa = 7.81QHH80 pKa = 6.85DD81 pKa = 4.36LKK83 pKa = 11.26CGGEE87 pKa = 4.04RR88 pKa = 11.84EE89 pKa = 4.03QLLFSRR95 pKa = 11.84MKK97 pKa = 10.51VDD99 pKa = 4.16QLLLANSKK107 pKa = 9.72GRR109 pKa = 11.84APQMVKK115 pKa = 10.93DD116 pKa = 4.0NLLSGVLNEE125 pKa = 4.08IKK127 pKa = 10.52KK128 pKa = 10.13RR129 pKa = 11.84VPKK132 pKa = 10.77SEE134 pKa = 4.04DD135 pKa = 3.07RR136 pKa = 11.84CTLGDD141 pKa = 3.9SDD143 pKa = 5.74LKK145 pKa = 10.56TMLNGFEE152 pKa = 4.43KK153 pKa = 11.1VNAGLNNPQRR163 pKa = 11.84LEE165 pKa = 3.82SWDD168 pKa = 3.48PSGTVDD174 pKa = 5.27LGRR177 pKa = 11.84LSLLGNPQRR186 pKa = 11.84LGWRR190 pKa = 11.84LGHH193 pKa = 7.2LKK195 pKa = 10.67ALTCAQLVILLDD207 pKa = 3.9FLVKK211 pKa = 10.59ALTDD215 pKa = 3.57MEE217 pKa = 4.6EE218 pKa = 4.18EE219 pKa = 4.42GSEE222 pKa = 4.64GEE224 pKa = 4.13GSLWGARR231 pKa = 11.84HH232 pKa = 5.1EE233 pKa = 4.37HH234 pKa = 6.25SSRR237 pKa = 11.84GCRR240 pKa = 11.84YY241 pKa = 9.11YY242 pKa = 11.01KK243 pKa = 10.51SGMCFKK249 pKa = 10.21MGRR252 pKa = 11.84CLYY255 pKa = 10.33SHH257 pKa = 6.92NPYY260 pKa = 10.8KK261 pKa = 10.83LGASDD266 pKa = 5.52LIRR269 pKa = 11.84GIAIGLQSLDD279 pKa = 4.37LGSQPLCEE287 pKa = 4.4DD288 pKa = 3.09SGMPSLPQDD297 pKa = 3.04RR298 pKa = 11.84KK299 pKa = 10.28ARR301 pKa = 11.84KK302 pKa = 9.69DD303 pKa = 3.15MARR306 pKa = 11.84ALISEE311 pKa = 4.41SAINPRR317 pKa = 11.84LPKK320 pKa = 10.44EE321 pKa = 4.26SLLTMAQLIWEE332 pKa = 4.36WDD334 pKa = 3.56YY335 pKa = 11.88VPPSEE340 pKa = 5.53SGPTPPSSGDD350 pKa = 3.23VPGEE354 pKa = 3.93VRR356 pKa = 11.84DD357 pKa = 3.77MLLTLKK363 pKa = 10.73ALEE366 pKa = 4.52GPKK369 pKa = 10.4GVAAALTRR377 pKa = 11.84YY378 pKa = 8.72SQEE381 pKa = 3.32HH382 pKa = 6.69RR383 pKa = 11.84GRR385 pKa = 11.84LLISMEE391 pKa = 3.99KK392 pKa = 10.6KK393 pKa = 8.95MM394 pKa = 4.99
Molecular weight: 43.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4352 |
221 |
2146 |
725.3 |
80.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.043 ± 0.899 | 2.022 ± 0.264 |
5.331 ± 0.651 | 5.767 ± 0.723 |
3.699 ± 0.454 | 7.835 ± 0.759 |
2.642 ± 0.604 | 4.848 ± 0.483 |
4.894 ± 0.96 | 11.213 ± 0.795 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.24 ± 0.504 | 3.608 ± 0.446 |
4.825 ± 0.299 | 3.378 ± 0.221 |
6.296 ± 0.349 | 7.813 ± 0.818 |
6.319 ± 0.511 | 6.664 ± 0.761 |
1.241 ± 0.082 | 2.321 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
