
Tortoise microvirus 77
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
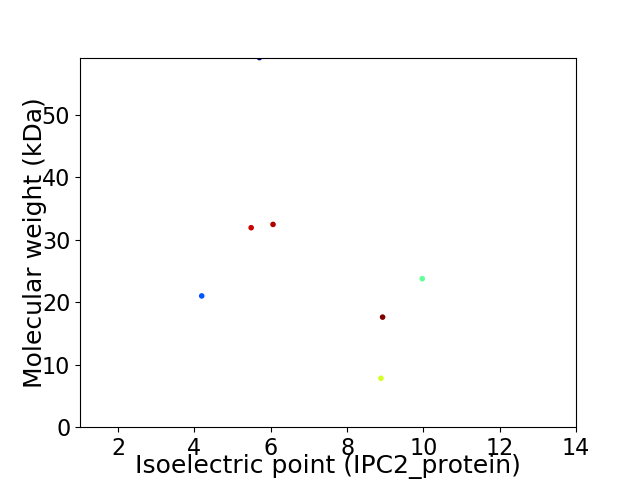
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
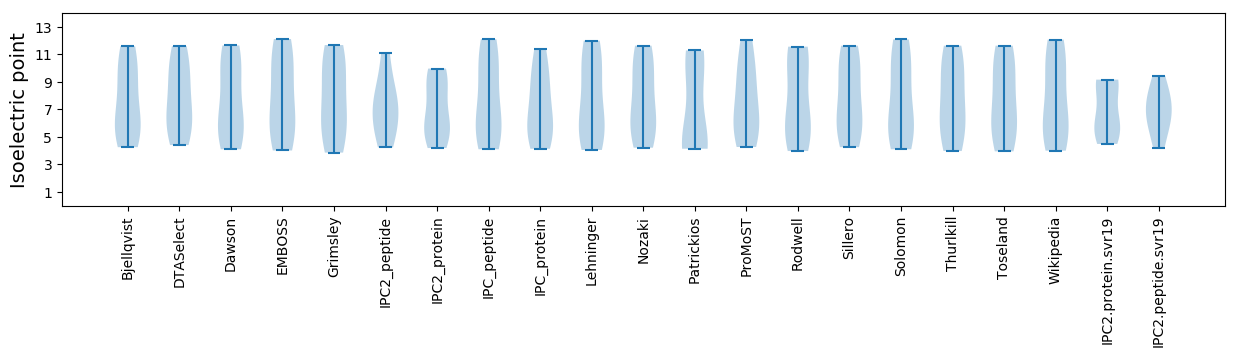
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6V8|A0A4P8W6V8_9VIRU Uncharacterized protein OS=Tortoise microvirus 77 OX=2583184 PE=4 SV=1
MM1 pKa = 7.64AIKK4 pKa = 10.69VGDD7 pKa = 3.66LNRR10 pKa = 11.84WKK12 pKa = 10.49KK13 pKa = 10.54LKK15 pKa = 10.46PGGALEE21 pKa = 4.53LRR23 pKa = 11.84SDD25 pKa = 3.43KK26 pKa = 10.47QRR28 pKa = 11.84QVRR31 pKa = 11.84LEE33 pKa = 4.19VNVDD37 pKa = 3.14QPTQVMALRR46 pKa = 11.84GTDD49 pKa = 3.53LTLLGVVMGMDD60 pKa = 3.85VIEE63 pKa = 4.67FVTDD67 pKa = 4.02GPTEE71 pKa = 4.24VVFSTEE77 pKa = 3.84GEE79 pKa = 4.21VFYY82 pKa = 8.06FTRR85 pKa = 11.84DD86 pKa = 3.26GDD88 pKa = 4.46SEE90 pKa = 4.76ALDD93 pKa = 3.82LLEE96 pKa = 4.49EE97 pKa = 4.29VSFATLANRR106 pKa = 11.84RR107 pKa = 11.84ARR109 pKa = 11.84NPEE112 pKa = 3.68MEE114 pKa = 4.67LMFFKK119 pKa = 10.4MNQNIEE125 pKa = 3.92RR126 pKa = 11.84RR127 pKa = 11.84LAIQADD133 pKa = 3.84DD134 pKa = 3.78FEE136 pKa = 6.05RR137 pKa = 11.84RR138 pKa = 11.84LQEE141 pKa = 3.7MGYY144 pKa = 10.63DD145 pKa = 3.73PDD147 pKa = 3.78TGEE150 pKa = 5.07VSDD153 pKa = 5.67ADD155 pKa = 3.83DD156 pKa = 3.74TGTAGGDD163 pKa = 3.24TGEE166 pKa = 4.97AEE168 pKa = 4.23AEE170 pKa = 4.14SGGVGGADD178 pKa = 3.15AEE180 pKa = 4.56APGDD184 pKa = 3.71AASAPGGGAGTAA196 pKa = 3.64
MM1 pKa = 7.64AIKK4 pKa = 10.69VGDD7 pKa = 3.66LNRR10 pKa = 11.84WKK12 pKa = 10.49KK13 pKa = 10.54LKK15 pKa = 10.46PGGALEE21 pKa = 4.53LRR23 pKa = 11.84SDD25 pKa = 3.43KK26 pKa = 10.47QRR28 pKa = 11.84QVRR31 pKa = 11.84LEE33 pKa = 4.19VNVDD37 pKa = 3.14QPTQVMALRR46 pKa = 11.84GTDD49 pKa = 3.53LTLLGVVMGMDD60 pKa = 3.85VIEE63 pKa = 4.67FVTDD67 pKa = 4.02GPTEE71 pKa = 4.24VVFSTEE77 pKa = 3.84GEE79 pKa = 4.21VFYY82 pKa = 8.06FTRR85 pKa = 11.84DD86 pKa = 3.26GDD88 pKa = 4.46SEE90 pKa = 4.76ALDD93 pKa = 3.82LLEE96 pKa = 4.49EE97 pKa = 4.29VSFATLANRR106 pKa = 11.84RR107 pKa = 11.84ARR109 pKa = 11.84NPEE112 pKa = 3.68MEE114 pKa = 4.67LMFFKK119 pKa = 10.4MNQNIEE125 pKa = 3.92RR126 pKa = 11.84RR127 pKa = 11.84LAIQADD133 pKa = 3.84DD134 pKa = 3.78FEE136 pKa = 6.05RR137 pKa = 11.84RR138 pKa = 11.84LQEE141 pKa = 3.7MGYY144 pKa = 10.63DD145 pKa = 3.73PDD147 pKa = 3.78TGEE150 pKa = 5.07VSDD153 pKa = 5.67ADD155 pKa = 3.83DD156 pKa = 3.74TGTAGGDD163 pKa = 3.24TGEE166 pKa = 4.97AEE168 pKa = 4.23AEE170 pKa = 4.14SGGVGGADD178 pKa = 3.15AEE180 pKa = 4.56APGDD184 pKa = 3.71AASAPGGGAGTAA196 pKa = 3.64
Molecular weight: 20.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6K2|A0A4P8W6K2_9VIRU Peptidase_M15_4 domain-containing protein OS=Tortoise microvirus 77 OX=2583184 PE=4 SV=1
MM1 pKa = 7.19QSVAKK6 pKa = 10.25DD7 pKa = 3.27SRR9 pKa = 11.84EE10 pKa = 3.76NPGRR14 pKa = 11.84NRR16 pKa = 11.84ISTSKK21 pKa = 10.4IGGQTTGAPRR31 pKa = 11.84KK32 pKa = 5.62TTAAAPAKK40 pKa = 10.11QSAPARR46 pKa = 11.84KK47 pKa = 8.02ATPKK51 pKa = 10.25AAPDD55 pKa = 4.71PIRR58 pKa = 11.84RR59 pKa = 11.84ATQAAPSRR67 pKa = 11.84ATLEE71 pKa = 4.17RR72 pKa = 11.84QAGLPAQPARR82 pKa = 11.84RR83 pKa = 11.84SVPSHH88 pKa = 6.57SSSEE92 pKa = 4.2SLSGSEE98 pKa = 4.74GGEE101 pKa = 4.01TTLGRR106 pKa = 11.84TARR109 pKa = 11.84EE110 pKa = 3.62RR111 pKa = 11.84LAQQAVQPPRR121 pKa = 11.84APDD124 pKa = 3.71PVILAPTNPITPVNPVQEE142 pKa = 4.17AARR145 pKa = 11.84ARR147 pKa = 11.84RR148 pKa = 11.84VHH150 pKa = 6.14SLPGGTLRR158 pKa = 11.84GTQNNTTRR166 pKa = 11.84TVTAKK171 pKa = 10.6AKK173 pKa = 10.56PGDD176 pKa = 3.62AKK178 pKa = 11.04ARR180 pKa = 11.84TQVQPHH186 pKa = 6.15KK187 pKa = 10.43AARR190 pKa = 11.84DD191 pKa = 3.42DD192 pKa = 3.9RR193 pKa = 11.84EE194 pKa = 3.65PDD196 pKa = 3.05KK197 pKa = 11.41RR198 pKa = 11.84LVCKK202 pKa = 10.46ARR204 pKa = 11.84PEE206 pKa = 4.32SNKK209 pKa = 10.04PKK211 pKa = 10.61GGGGGAKK218 pKa = 9.72RR219 pKa = 11.84AFIPWCSS226 pKa = 3.07
MM1 pKa = 7.19QSVAKK6 pKa = 10.25DD7 pKa = 3.27SRR9 pKa = 11.84EE10 pKa = 3.76NPGRR14 pKa = 11.84NRR16 pKa = 11.84ISTSKK21 pKa = 10.4IGGQTTGAPRR31 pKa = 11.84KK32 pKa = 5.62TTAAAPAKK40 pKa = 10.11QSAPARR46 pKa = 11.84KK47 pKa = 8.02ATPKK51 pKa = 10.25AAPDD55 pKa = 4.71PIRR58 pKa = 11.84RR59 pKa = 11.84ATQAAPSRR67 pKa = 11.84ATLEE71 pKa = 4.17RR72 pKa = 11.84QAGLPAQPARR82 pKa = 11.84RR83 pKa = 11.84SVPSHH88 pKa = 6.57SSSEE92 pKa = 4.2SLSGSEE98 pKa = 4.74GGEE101 pKa = 4.01TTLGRR106 pKa = 11.84TARR109 pKa = 11.84EE110 pKa = 3.62RR111 pKa = 11.84LAQQAVQPPRR121 pKa = 11.84APDD124 pKa = 3.71PVILAPTNPITPVNPVQEE142 pKa = 4.17AARR145 pKa = 11.84ARR147 pKa = 11.84RR148 pKa = 11.84VHH150 pKa = 6.14SLPGGTLRR158 pKa = 11.84GTQNNTTRR166 pKa = 11.84TVTAKK171 pKa = 10.6AKK173 pKa = 10.56PGDD176 pKa = 3.62AKK178 pKa = 11.04ARR180 pKa = 11.84TQVQPHH186 pKa = 6.15KK187 pKa = 10.43AARR190 pKa = 11.84DD191 pKa = 3.42DD192 pKa = 3.9RR193 pKa = 11.84EE194 pKa = 3.65PDD196 pKa = 3.05KK197 pKa = 11.41RR198 pKa = 11.84LVCKK202 pKa = 10.46ARR204 pKa = 11.84PEE206 pKa = 4.32SNKK209 pKa = 10.04PKK211 pKa = 10.61GGGGGAKK218 pKa = 9.72RR219 pKa = 11.84AFIPWCSS226 pKa = 3.07
Molecular weight: 23.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1769 |
74 |
537 |
252.7 |
27.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.91 ± 1.115 | 0.17 ± 0.139 |
5.596 ± 0.658 | 6.162 ± 0.995 |
3.222 ± 0.516 | 9.723 ± 1.052 |
1.922 ± 0.604 | 3.166 ± 0.289 |
4.409 ± 0.584 | 7.349 ± 0.811 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.445 | 3.505 ± 0.765 |
6.897 ± 1.059 | 4.635 ± 0.399 |
7.405 ± 0.995 | 5.427 ± 0.622 |
5.709 ± 0.805 | 7.01 ± 1.248 |
2.374 ± 0.664 | 2.261 ± 0.643 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
