
Trichoplusia ni TED virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Metaviridae; Errantivirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
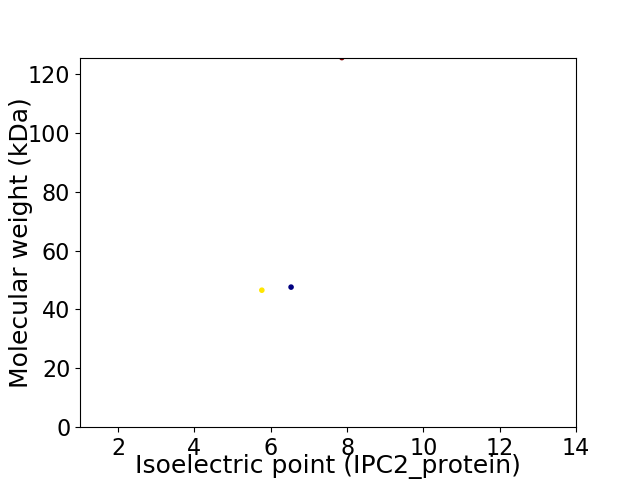
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
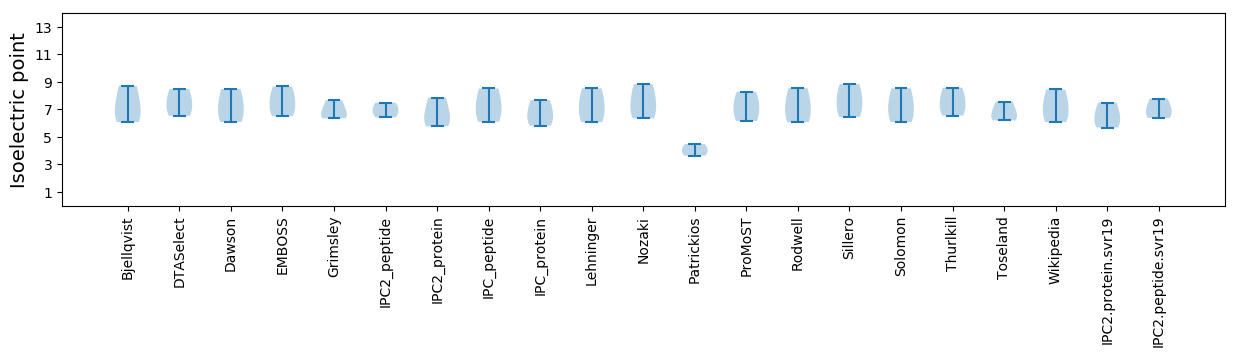
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65353|Q65353_9VIRU ORF B OS=Trichoplusia ni TED virus OX=2083181 PE=4 SV=1
MM1 pKa = 7.47PHH3 pKa = 7.06DD4 pKa = 4.68PDD6 pKa = 4.61VIFKK10 pKa = 10.68ALRR13 pKa = 11.84LVPEE17 pKa = 4.49FNGNPNILTRR27 pKa = 11.84FINICDD33 pKa = 3.8KK34 pKa = 11.05LVEE37 pKa = 4.34QYY39 pKa = 11.57ASAEE43 pKa = 4.13PGSEE47 pKa = 4.21LGNLCLLNGILNKK60 pKa = 9.29VTGTAASTINANGIPEE76 pKa = 4.21TWVGIRR82 pKa = 11.84SSLINNFSDD91 pKa = 4.42QRR93 pKa = 11.84DD94 pKa = 3.53EE95 pKa = 4.04TALYY99 pKa = 10.68NDD101 pKa = 5.19LSLASQGNKK110 pKa = 8.47TPQEE114 pKa = 4.35FYY116 pKa = 10.41EE117 pKa = 4.16QCQTLFSTIMTYY129 pKa = 8.58VTLHH133 pKa = 5.63EE134 pKa = 4.46TLPTTIEE141 pKa = 4.06AKK143 pKa = 9.59RR144 pKa = 11.84ALYY147 pKa = 10.42KK148 pKa = 10.51KK149 pKa = 9.0VTVQAFVRR157 pKa = 11.84GLKK160 pKa = 10.12EE161 pKa = 3.74PLGSRR166 pKa = 11.84IRR168 pKa = 11.84CMRR171 pKa = 11.84PEE173 pKa = 4.37TIEE176 pKa = 3.86KK177 pKa = 10.03ALEE180 pKa = 3.99YY181 pKa = 10.46VQEE184 pKa = 4.2EE185 pKa = 4.66LNVIYY190 pKa = 9.95LQQRR194 pKa = 11.84NEE196 pKa = 3.74SSRR199 pKa = 11.84AHH201 pKa = 6.7SSPKK205 pKa = 9.52MLPIPQQSPVTPFNTLGIHH224 pKa = 6.76RR225 pKa = 11.84PPVPNWPVPMGQRR238 pKa = 11.84GNQPPPQPFKK248 pKa = 11.34FNVPNQYY255 pKa = 10.36HH256 pKa = 5.86NRR258 pKa = 11.84MPTKK262 pKa = 8.12TQQMLRR268 pKa = 11.84APPPNYY274 pKa = 9.69HH275 pKa = 6.19PQSNVFRR282 pKa = 11.84LPPRR286 pKa = 11.84NPPPNQIVKK295 pKa = 9.97PMSGVQHH302 pKa = 6.26FVPKK306 pKa = 9.31TLPVMTGHH314 pKa = 7.7DD315 pKa = 3.51WRR317 pKa = 11.84KK318 pKa = 10.37SGNPPPNNYY327 pKa = 9.75FKK329 pKa = 10.65TRR331 pKa = 11.84EE332 pKa = 3.98LNVNEE337 pKa = 5.34FYY339 pKa = 11.26SSDD342 pKa = 3.44DD343 pKa = 3.54SYY345 pKa = 12.24NSVDD349 pKa = 3.92YY350 pKa = 11.01YY351 pKa = 11.25SEE353 pKa = 4.71PGCDD357 pKa = 3.54YY358 pKa = 10.76YY359 pKa = 11.18TDD361 pKa = 4.42YY362 pKa = 11.73YY363 pKa = 10.46NNPYY367 pKa = 10.73DD368 pKa = 4.03YY369 pKa = 9.21DD370 pKa = 4.21TNATCYY376 pKa = 10.24DD377 pKa = 3.86LPYY380 pKa = 10.47DD381 pKa = 3.9ASEE384 pKa = 4.61TEE386 pKa = 4.15AQPGPSHH393 pKa = 5.49VHH395 pKa = 6.26EE396 pKa = 4.72SQDD399 pKa = 3.62FQSTKK404 pKa = 10.22PSNEE408 pKa = 3.72QGG410 pKa = 3.25
MM1 pKa = 7.47PHH3 pKa = 7.06DD4 pKa = 4.68PDD6 pKa = 4.61VIFKK10 pKa = 10.68ALRR13 pKa = 11.84LVPEE17 pKa = 4.49FNGNPNILTRR27 pKa = 11.84FINICDD33 pKa = 3.8KK34 pKa = 11.05LVEE37 pKa = 4.34QYY39 pKa = 11.57ASAEE43 pKa = 4.13PGSEE47 pKa = 4.21LGNLCLLNGILNKK60 pKa = 9.29VTGTAASTINANGIPEE76 pKa = 4.21TWVGIRR82 pKa = 11.84SSLINNFSDD91 pKa = 4.42QRR93 pKa = 11.84DD94 pKa = 3.53EE95 pKa = 4.04TALYY99 pKa = 10.68NDD101 pKa = 5.19LSLASQGNKK110 pKa = 8.47TPQEE114 pKa = 4.35FYY116 pKa = 10.41EE117 pKa = 4.16QCQTLFSTIMTYY129 pKa = 8.58VTLHH133 pKa = 5.63EE134 pKa = 4.46TLPTTIEE141 pKa = 4.06AKK143 pKa = 9.59RR144 pKa = 11.84ALYY147 pKa = 10.42KK148 pKa = 10.51KK149 pKa = 9.0VTVQAFVRR157 pKa = 11.84GLKK160 pKa = 10.12EE161 pKa = 3.74PLGSRR166 pKa = 11.84IRR168 pKa = 11.84CMRR171 pKa = 11.84PEE173 pKa = 4.37TIEE176 pKa = 3.86KK177 pKa = 10.03ALEE180 pKa = 3.99YY181 pKa = 10.46VQEE184 pKa = 4.2EE185 pKa = 4.66LNVIYY190 pKa = 9.95LQQRR194 pKa = 11.84NEE196 pKa = 3.74SSRR199 pKa = 11.84AHH201 pKa = 6.7SSPKK205 pKa = 9.52MLPIPQQSPVTPFNTLGIHH224 pKa = 6.76RR225 pKa = 11.84PPVPNWPVPMGQRR238 pKa = 11.84GNQPPPQPFKK248 pKa = 11.34FNVPNQYY255 pKa = 10.36HH256 pKa = 5.86NRR258 pKa = 11.84MPTKK262 pKa = 8.12TQQMLRR268 pKa = 11.84APPPNYY274 pKa = 9.69HH275 pKa = 6.19PQSNVFRR282 pKa = 11.84LPPRR286 pKa = 11.84NPPPNQIVKK295 pKa = 9.97PMSGVQHH302 pKa = 6.26FVPKK306 pKa = 9.31TLPVMTGHH314 pKa = 7.7DD315 pKa = 3.51WRR317 pKa = 11.84KK318 pKa = 10.37SGNPPPNNYY327 pKa = 9.75FKK329 pKa = 10.65TRR331 pKa = 11.84EE332 pKa = 3.98LNVNEE337 pKa = 5.34FYY339 pKa = 11.26SSDD342 pKa = 3.44DD343 pKa = 3.54SYY345 pKa = 12.24NSVDD349 pKa = 3.92YY350 pKa = 11.01YY351 pKa = 11.25SEE353 pKa = 4.71PGCDD357 pKa = 3.54YY358 pKa = 10.76YY359 pKa = 11.18TDD361 pKa = 4.42YY362 pKa = 11.73YY363 pKa = 10.46NNPYY367 pKa = 10.73DD368 pKa = 4.03YY369 pKa = 9.21DD370 pKa = 4.21TNATCYY376 pKa = 10.24DD377 pKa = 3.86LPYY380 pKa = 10.47DD381 pKa = 3.9ASEE384 pKa = 4.61TEE386 pKa = 4.15AQPGPSHH393 pKa = 5.49VHH395 pKa = 6.26EE396 pKa = 4.72SQDD399 pKa = 3.62FQSTKK404 pKa = 10.22PSNEE408 pKa = 3.72QGG410 pKa = 3.25
Molecular weight: 46.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65354|Q65354_9VIRU ORF C OS=Trichoplusia ni TED virus OX=2083181 PE=4 SV=1
MM1 pKa = 7.38IPARR5 pKa = 11.84SSKK8 pKa = 10.3LVRR11 pKa = 11.84IPINATDD18 pKa = 4.05GEE20 pKa = 4.73VLVEE24 pKa = 3.92EE25 pKa = 5.29QMFCNCIVHH34 pKa = 6.23EE35 pKa = 4.53CVTMVKK41 pKa = 10.25DD42 pKa = 3.29GRR44 pKa = 11.84GYY46 pKa = 11.08VEE48 pKa = 5.54LEE50 pKa = 3.89NPTPNDD56 pKa = 3.1VIFYY60 pKa = 10.13LDD62 pKa = 3.39QPASAEE68 pKa = 4.03LFNIKK73 pKa = 8.53CTQVEE78 pKa = 3.93QSQRR82 pKa = 11.84VDD84 pKa = 3.31DD85 pKa = 3.94VLSRR89 pKa = 11.84LRR91 pKa = 11.84TDD93 pKa = 3.51HH94 pKa = 7.12LNEE97 pKa = 3.92EE98 pKa = 4.41EE99 pKa = 4.27KK100 pKa = 11.36ANLLRR105 pKa = 11.84LCSRR109 pKa = 11.84YY110 pKa = 10.27SDD112 pKa = 3.41VFYY115 pKa = 10.69IDD117 pKa = 4.92GEE119 pKa = 4.35ALTFTNKK126 pKa = 9.08IKK128 pKa = 10.64HH129 pKa = 6.57RR130 pKa = 11.84IRR132 pKa = 11.84TTDD135 pKa = 3.38EE136 pKa = 3.92VPVYY140 pKa = 8.82TKK142 pKa = 10.26SYY144 pKa = 9.48RR145 pKa = 11.84YY146 pKa = 9.55PFIHH150 pKa = 6.07RR151 pKa = 11.84QEE153 pKa = 4.06VRR155 pKa = 11.84DD156 pKa = 4.2QITKK160 pKa = 9.53MLDD163 pKa = 2.92QGIIRR168 pKa = 11.84PSDD171 pKa = 3.79SAWSSPIWVVPKK183 pKa = 10.67KK184 pKa = 10.04IDD186 pKa = 3.79ASGKK190 pKa = 7.82QKK192 pKa = 9.69WRR194 pKa = 11.84LVVDD198 pKa = 4.37FRR200 pKa = 11.84KK201 pKa = 10.49LNEE204 pKa = 3.68KK205 pKa = 9.8TIDD208 pKa = 3.73DD209 pKa = 4.9KK210 pKa = 11.96YY211 pKa = 10.26PIPNISDD218 pKa = 3.49VLDD221 pKa = 3.72KK222 pKa = 11.16LGKK225 pKa = 9.22CQYY228 pKa = 8.23FTTLDD233 pKa = 3.85LASGFYY239 pKa = 9.36QVEE242 pKa = 4.36MDD244 pKa = 4.09PQDD247 pKa = 3.14ISKK250 pKa = 8.82TAFNVEE256 pKa = 3.98HH257 pKa = 6.24GHH259 pKa = 6.73FEE261 pKa = 4.25FLRR264 pKa = 11.84MPMGLKK270 pKa = 10.23NSPSTFQRR278 pKa = 11.84VMDD281 pKa = 3.83NVLRR285 pKa = 11.84GLQNNICLVYY295 pKa = 10.53LDD297 pKa = 5.49DD298 pKa = 5.52IIVYY302 pKa = 7.98STSLQEE308 pKa = 3.93HH309 pKa = 6.83LEE311 pKa = 3.8NLEE314 pKa = 4.22RR315 pKa = 11.84VFQRR319 pKa = 11.84LRR321 pKa = 11.84EE322 pKa = 4.32SNFKK326 pKa = 10.37IQMDD330 pKa = 3.23KK331 pKa = 11.47SEE333 pKa = 4.08FLKK336 pKa = 11.32LEE338 pKa = 3.97TAYY341 pKa = 10.56LGHH344 pKa = 7.09IISRR348 pKa = 11.84DD349 pKa = 3.71GIKK352 pKa = 10.47PNPDD356 pKa = 3.82KK357 pKa = 10.97ISAIQKK363 pKa = 8.88YY364 pKa = 9.82LIPKK368 pKa = 7.35TPKK371 pKa = 9.95EE372 pKa = 3.84IKK374 pKa = 10.23QFLGLLGYY382 pKa = 8.7YY383 pKa = 10.12RR384 pKa = 11.84KK385 pKa = 10.1FIPDD389 pKa = 4.22FARR392 pKa = 11.84LTKK395 pKa = 10.38PLTQCLKK402 pKa = 10.36KK403 pKa = 10.5GSKK406 pKa = 8.61VTLSPEE412 pKa = 3.78YY413 pKa = 10.82VNAFEE418 pKa = 4.5HH419 pKa = 6.85CKK421 pKa = 9.5TLLTNDD427 pKa = 5.68PILQYY432 pKa = 11.01PDD434 pKa = 3.31FTRR437 pKa = 11.84EE438 pKa = 3.84FNLTTDD444 pKa = 3.7ASNFAIGAVLSQGPIGSDD462 pKa = 3.06KK463 pKa = 10.15PVCYY467 pKa = 10.28ASRR470 pKa = 11.84TLNEE474 pKa = 3.98SEE476 pKa = 4.93LNYY479 pKa = 10.11STIEE483 pKa = 4.03KK484 pKa = 9.98EE485 pKa = 3.78LLAIVWATKK494 pKa = 8.79YY495 pKa = 9.54FRR497 pKa = 11.84PYY499 pKa = 10.58LFGRR503 pKa = 11.84KK504 pKa = 8.29FKK506 pKa = 10.68ILTDD510 pKa = 3.85HH511 pKa = 7.48KK512 pKa = 10.1PLQWMMNLKK521 pKa = 10.37DD522 pKa = 3.71PNSRR526 pKa = 11.84MTRR529 pKa = 11.84WRR531 pKa = 11.84LRR533 pKa = 11.84LSEE536 pKa = 4.09YY537 pKa = 10.83DD538 pKa = 3.36FSVVYY543 pKa = 10.5KK544 pKa = 10.44KK545 pKa = 11.03GKK547 pKa = 10.39SNTNADD553 pKa = 3.32ALSRR557 pKa = 11.84VEE559 pKa = 3.78IHH561 pKa = 5.08TTEE564 pKa = 3.63IDD566 pKa = 3.86EE567 pKa = 4.32IDD569 pKa = 3.89SVIEE573 pKa = 4.14NIKK576 pKa = 10.15EE577 pKa = 3.83LSSMINNPSEE587 pKa = 3.78TSRR590 pKa = 11.84PQRR593 pKa = 11.84QTEE596 pKa = 4.01NTDD599 pKa = 3.55EE600 pKa = 3.99IRR602 pKa = 11.84QNSTTDD608 pKa = 3.23TVHH611 pKa = 6.7TSDD614 pKa = 3.51EE615 pKa = 4.35HH616 pKa = 6.86PILEE620 pKa = 4.24VPITNEE626 pKa = 3.36PLNRR630 pKa = 11.84FHH632 pKa = 7.05RR633 pKa = 11.84QIHH636 pKa = 5.09LTVVGDD642 pKa = 3.61IKK644 pKa = 10.69RR645 pKa = 11.84RR646 pKa = 11.84PIVTKK651 pKa = 9.73PFEE654 pKa = 4.1SHH656 pKa = 4.55TRR658 pKa = 11.84IAIQLSEE665 pKa = 4.67SNLEE669 pKa = 3.67QDD671 pKa = 3.84VISAIKK677 pKa = 10.1EE678 pKa = 3.98YY679 pKa = 11.26VNPKK683 pKa = 10.0VKK685 pKa = 9.34TALIINPPLKK695 pKa = 9.3MYY697 pKa = 10.4SIIPIIQKK705 pKa = 7.24TFRR708 pKa = 11.84SSSLNLVLTKK718 pKa = 10.65VEE720 pKa = 4.09LEE722 pKa = 4.05NVKK725 pKa = 10.45EE726 pKa = 3.98YY727 pKa = 10.95LRR729 pKa = 11.84QQDD732 pKa = 4.15IIRR735 pKa = 11.84HH736 pKa = 4.3YY737 pKa = 11.13HH738 pKa = 6.3DD739 pKa = 4.28GKK741 pKa = 9.18TNHH744 pKa = 6.77RR745 pKa = 11.84GINEE749 pKa = 4.29CYY751 pKa = 9.49LALSKK756 pKa = 10.42RR757 pKa = 11.84YY758 pKa = 8.28YY759 pKa = 8.04WPRR762 pKa = 11.84MKK764 pKa = 10.75DD765 pKa = 3.21QITKK769 pKa = 10.02FINEE773 pKa = 4.21CTICGQAKK781 pKa = 9.97YY782 pKa = 10.1DD783 pKa = 3.87RR784 pKa = 11.84NPIRR788 pKa = 11.84PQFNIVPPATKK799 pKa = 9.91PLEE802 pKa = 4.33TVHH805 pKa = 6.58MDD807 pKa = 3.82LFTVQNEE814 pKa = 4.24KK815 pKa = 11.02YY816 pKa = 8.74ITFIDD821 pKa = 3.79VFTKK825 pKa = 10.67YY826 pKa = 10.26GQAYY830 pKa = 9.37HH831 pKa = 6.72LRR833 pKa = 11.84DD834 pKa = 3.34GTAISILQALLRR846 pKa = 11.84FCTHH850 pKa = 6.68HH851 pKa = 6.7GLPITIVTDD860 pKa = 3.12NGTEE864 pKa = 3.95FSNQLFSEE872 pKa = 5.06FVRR875 pKa = 11.84IHH877 pKa = 6.61KK878 pKa = 9.97IIHH881 pKa = 6.41HH882 pKa = 6.05KK883 pKa = 8.7TLPHH887 pKa = 6.26SPSDD891 pKa = 3.25NGNIEE896 pKa = 4.37RR897 pKa = 11.84FHH899 pKa = 5.67STILEE904 pKa = 4.36HH905 pKa = 8.01IRR907 pKa = 11.84ILKK910 pKa = 8.99LQHH913 pKa = 6.37KK914 pKa = 9.76DD915 pKa = 3.41EE916 pKa = 5.77PIVNLMPYY924 pKa = 10.25AIIGYY929 pKa = 9.07NSSIHH934 pKa = 6.66SFTKK938 pKa = 10.26CRR940 pKa = 11.84PFDD943 pKa = 3.9LLNGHH948 pKa = 6.76FDD950 pKa = 3.54PRR952 pKa = 11.84DD953 pKa = 3.62PLDD956 pKa = 3.81IDD958 pKa = 3.7LTEE961 pKa = 5.91HH962 pKa = 7.57ILQQYY967 pKa = 8.17AQNHH971 pKa = 4.75RR972 pKa = 11.84QQMKK976 pKa = 9.39QVYY979 pKa = 9.08EE980 pKa = 4.42IINEE984 pKa = 3.93TSLANRR990 pKa = 11.84TALIEE995 pKa = 4.3SRR997 pKa = 11.84NKK999 pKa = 8.59TRR1001 pKa = 11.84EE1002 pKa = 3.98SEE1004 pKa = 4.15VEE1006 pKa = 4.26YY1007 pKa = 10.49IPQQQVFIKK1016 pKa = 10.5NPLASRR1022 pKa = 11.84QKK1024 pKa = 9.15VAPRR1028 pKa = 11.84YY1029 pKa = 6.94TQDD1032 pKa = 2.87TVLADD1037 pKa = 4.19LPIHH1041 pKa = 7.03IYY1043 pKa = 8.5TSKK1046 pKa = 10.68KK1047 pKa = 9.3RR1048 pKa = 11.84GPVAKK1053 pKa = 10.21ARR1055 pKa = 11.84LKK1057 pKa = 10.42RR1058 pKa = 11.84VPKK1061 pKa = 10.72GNTLLQDD1068 pKa = 3.6SAATDD1073 pKa = 3.71NTCDD1077 pKa = 3.17ASSRR1081 pKa = 11.84DD1082 pKa = 3.55KK1083 pKa = 10.97TT1084 pKa = 3.75
MM1 pKa = 7.38IPARR5 pKa = 11.84SSKK8 pKa = 10.3LVRR11 pKa = 11.84IPINATDD18 pKa = 4.05GEE20 pKa = 4.73VLVEE24 pKa = 3.92EE25 pKa = 5.29QMFCNCIVHH34 pKa = 6.23EE35 pKa = 4.53CVTMVKK41 pKa = 10.25DD42 pKa = 3.29GRR44 pKa = 11.84GYY46 pKa = 11.08VEE48 pKa = 5.54LEE50 pKa = 3.89NPTPNDD56 pKa = 3.1VIFYY60 pKa = 10.13LDD62 pKa = 3.39QPASAEE68 pKa = 4.03LFNIKK73 pKa = 8.53CTQVEE78 pKa = 3.93QSQRR82 pKa = 11.84VDD84 pKa = 3.31DD85 pKa = 3.94VLSRR89 pKa = 11.84LRR91 pKa = 11.84TDD93 pKa = 3.51HH94 pKa = 7.12LNEE97 pKa = 3.92EE98 pKa = 4.41EE99 pKa = 4.27KK100 pKa = 11.36ANLLRR105 pKa = 11.84LCSRR109 pKa = 11.84YY110 pKa = 10.27SDD112 pKa = 3.41VFYY115 pKa = 10.69IDD117 pKa = 4.92GEE119 pKa = 4.35ALTFTNKK126 pKa = 9.08IKK128 pKa = 10.64HH129 pKa = 6.57RR130 pKa = 11.84IRR132 pKa = 11.84TTDD135 pKa = 3.38EE136 pKa = 3.92VPVYY140 pKa = 8.82TKK142 pKa = 10.26SYY144 pKa = 9.48RR145 pKa = 11.84YY146 pKa = 9.55PFIHH150 pKa = 6.07RR151 pKa = 11.84QEE153 pKa = 4.06VRR155 pKa = 11.84DD156 pKa = 4.2QITKK160 pKa = 9.53MLDD163 pKa = 2.92QGIIRR168 pKa = 11.84PSDD171 pKa = 3.79SAWSSPIWVVPKK183 pKa = 10.67KK184 pKa = 10.04IDD186 pKa = 3.79ASGKK190 pKa = 7.82QKK192 pKa = 9.69WRR194 pKa = 11.84LVVDD198 pKa = 4.37FRR200 pKa = 11.84KK201 pKa = 10.49LNEE204 pKa = 3.68KK205 pKa = 9.8TIDD208 pKa = 3.73DD209 pKa = 4.9KK210 pKa = 11.96YY211 pKa = 10.26PIPNISDD218 pKa = 3.49VLDD221 pKa = 3.72KK222 pKa = 11.16LGKK225 pKa = 9.22CQYY228 pKa = 8.23FTTLDD233 pKa = 3.85LASGFYY239 pKa = 9.36QVEE242 pKa = 4.36MDD244 pKa = 4.09PQDD247 pKa = 3.14ISKK250 pKa = 8.82TAFNVEE256 pKa = 3.98HH257 pKa = 6.24GHH259 pKa = 6.73FEE261 pKa = 4.25FLRR264 pKa = 11.84MPMGLKK270 pKa = 10.23NSPSTFQRR278 pKa = 11.84VMDD281 pKa = 3.83NVLRR285 pKa = 11.84GLQNNICLVYY295 pKa = 10.53LDD297 pKa = 5.49DD298 pKa = 5.52IIVYY302 pKa = 7.98STSLQEE308 pKa = 3.93HH309 pKa = 6.83LEE311 pKa = 3.8NLEE314 pKa = 4.22RR315 pKa = 11.84VFQRR319 pKa = 11.84LRR321 pKa = 11.84EE322 pKa = 4.32SNFKK326 pKa = 10.37IQMDD330 pKa = 3.23KK331 pKa = 11.47SEE333 pKa = 4.08FLKK336 pKa = 11.32LEE338 pKa = 3.97TAYY341 pKa = 10.56LGHH344 pKa = 7.09IISRR348 pKa = 11.84DD349 pKa = 3.71GIKK352 pKa = 10.47PNPDD356 pKa = 3.82KK357 pKa = 10.97ISAIQKK363 pKa = 8.88YY364 pKa = 9.82LIPKK368 pKa = 7.35TPKK371 pKa = 9.95EE372 pKa = 3.84IKK374 pKa = 10.23QFLGLLGYY382 pKa = 8.7YY383 pKa = 10.12RR384 pKa = 11.84KK385 pKa = 10.1FIPDD389 pKa = 4.22FARR392 pKa = 11.84LTKK395 pKa = 10.38PLTQCLKK402 pKa = 10.36KK403 pKa = 10.5GSKK406 pKa = 8.61VTLSPEE412 pKa = 3.78YY413 pKa = 10.82VNAFEE418 pKa = 4.5HH419 pKa = 6.85CKK421 pKa = 9.5TLLTNDD427 pKa = 5.68PILQYY432 pKa = 11.01PDD434 pKa = 3.31FTRR437 pKa = 11.84EE438 pKa = 3.84FNLTTDD444 pKa = 3.7ASNFAIGAVLSQGPIGSDD462 pKa = 3.06KK463 pKa = 10.15PVCYY467 pKa = 10.28ASRR470 pKa = 11.84TLNEE474 pKa = 3.98SEE476 pKa = 4.93LNYY479 pKa = 10.11STIEE483 pKa = 4.03KK484 pKa = 9.98EE485 pKa = 3.78LLAIVWATKK494 pKa = 8.79YY495 pKa = 9.54FRR497 pKa = 11.84PYY499 pKa = 10.58LFGRR503 pKa = 11.84KK504 pKa = 8.29FKK506 pKa = 10.68ILTDD510 pKa = 3.85HH511 pKa = 7.48KK512 pKa = 10.1PLQWMMNLKK521 pKa = 10.37DD522 pKa = 3.71PNSRR526 pKa = 11.84MTRR529 pKa = 11.84WRR531 pKa = 11.84LRR533 pKa = 11.84LSEE536 pKa = 4.09YY537 pKa = 10.83DD538 pKa = 3.36FSVVYY543 pKa = 10.5KK544 pKa = 10.44KK545 pKa = 11.03GKK547 pKa = 10.39SNTNADD553 pKa = 3.32ALSRR557 pKa = 11.84VEE559 pKa = 3.78IHH561 pKa = 5.08TTEE564 pKa = 3.63IDD566 pKa = 3.86EE567 pKa = 4.32IDD569 pKa = 3.89SVIEE573 pKa = 4.14NIKK576 pKa = 10.15EE577 pKa = 3.83LSSMINNPSEE587 pKa = 3.78TSRR590 pKa = 11.84PQRR593 pKa = 11.84QTEE596 pKa = 4.01NTDD599 pKa = 3.55EE600 pKa = 3.99IRR602 pKa = 11.84QNSTTDD608 pKa = 3.23TVHH611 pKa = 6.7TSDD614 pKa = 3.51EE615 pKa = 4.35HH616 pKa = 6.86PILEE620 pKa = 4.24VPITNEE626 pKa = 3.36PLNRR630 pKa = 11.84FHH632 pKa = 7.05RR633 pKa = 11.84QIHH636 pKa = 5.09LTVVGDD642 pKa = 3.61IKK644 pKa = 10.69RR645 pKa = 11.84RR646 pKa = 11.84PIVTKK651 pKa = 9.73PFEE654 pKa = 4.1SHH656 pKa = 4.55TRR658 pKa = 11.84IAIQLSEE665 pKa = 4.67SNLEE669 pKa = 3.67QDD671 pKa = 3.84VISAIKK677 pKa = 10.1EE678 pKa = 3.98YY679 pKa = 11.26VNPKK683 pKa = 10.0VKK685 pKa = 9.34TALIINPPLKK695 pKa = 9.3MYY697 pKa = 10.4SIIPIIQKK705 pKa = 7.24TFRR708 pKa = 11.84SSSLNLVLTKK718 pKa = 10.65VEE720 pKa = 4.09LEE722 pKa = 4.05NVKK725 pKa = 10.45EE726 pKa = 3.98YY727 pKa = 10.95LRR729 pKa = 11.84QQDD732 pKa = 4.15IIRR735 pKa = 11.84HH736 pKa = 4.3YY737 pKa = 11.13HH738 pKa = 6.3DD739 pKa = 4.28GKK741 pKa = 9.18TNHH744 pKa = 6.77RR745 pKa = 11.84GINEE749 pKa = 4.29CYY751 pKa = 9.49LALSKK756 pKa = 10.42RR757 pKa = 11.84YY758 pKa = 8.28YY759 pKa = 8.04WPRR762 pKa = 11.84MKK764 pKa = 10.75DD765 pKa = 3.21QITKK769 pKa = 10.02FINEE773 pKa = 4.21CTICGQAKK781 pKa = 9.97YY782 pKa = 10.1DD783 pKa = 3.87RR784 pKa = 11.84NPIRR788 pKa = 11.84PQFNIVPPATKK799 pKa = 9.91PLEE802 pKa = 4.33TVHH805 pKa = 6.58MDD807 pKa = 3.82LFTVQNEE814 pKa = 4.24KK815 pKa = 11.02YY816 pKa = 8.74ITFIDD821 pKa = 3.79VFTKK825 pKa = 10.67YY826 pKa = 10.26GQAYY830 pKa = 9.37HH831 pKa = 6.72LRR833 pKa = 11.84DD834 pKa = 3.34GTAISILQALLRR846 pKa = 11.84FCTHH850 pKa = 6.68HH851 pKa = 6.7GLPITIVTDD860 pKa = 3.12NGTEE864 pKa = 3.95FSNQLFSEE872 pKa = 5.06FVRR875 pKa = 11.84IHH877 pKa = 6.61KK878 pKa = 9.97IIHH881 pKa = 6.41HH882 pKa = 6.05KK883 pKa = 8.7TLPHH887 pKa = 6.26SPSDD891 pKa = 3.25NGNIEE896 pKa = 4.37RR897 pKa = 11.84FHH899 pKa = 5.67STILEE904 pKa = 4.36HH905 pKa = 8.01IRR907 pKa = 11.84ILKK910 pKa = 8.99LQHH913 pKa = 6.37KK914 pKa = 9.76DD915 pKa = 3.41EE916 pKa = 5.77PIVNLMPYY924 pKa = 10.25AIIGYY929 pKa = 9.07NSSIHH934 pKa = 6.66SFTKK938 pKa = 10.26CRR940 pKa = 11.84PFDD943 pKa = 3.9LLNGHH948 pKa = 6.76FDD950 pKa = 3.54PRR952 pKa = 11.84DD953 pKa = 3.62PLDD956 pKa = 3.81IDD958 pKa = 3.7LTEE961 pKa = 5.91HH962 pKa = 7.57ILQQYY967 pKa = 8.17AQNHH971 pKa = 4.75RR972 pKa = 11.84QQMKK976 pKa = 9.39QVYY979 pKa = 9.08EE980 pKa = 4.42IINEE984 pKa = 3.93TSLANRR990 pKa = 11.84TALIEE995 pKa = 4.3SRR997 pKa = 11.84NKK999 pKa = 8.59TRR1001 pKa = 11.84EE1002 pKa = 3.98SEE1004 pKa = 4.15VEE1006 pKa = 4.26YY1007 pKa = 10.49IPQQQVFIKK1016 pKa = 10.5NPLASRR1022 pKa = 11.84QKK1024 pKa = 9.15VAPRR1028 pKa = 11.84YY1029 pKa = 6.94TQDD1032 pKa = 2.87TVLADD1037 pKa = 4.19LPIHH1041 pKa = 7.03IYY1043 pKa = 8.5TSKK1046 pKa = 10.68KK1047 pKa = 9.3RR1048 pKa = 11.84GPVAKK1053 pKa = 10.21ARR1055 pKa = 11.84LKK1057 pKa = 10.42RR1058 pKa = 11.84VPKK1061 pKa = 10.72GNTLLQDD1068 pKa = 3.6SAATDD1073 pKa = 3.71NTCDD1077 pKa = 3.17ASSRR1081 pKa = 11.84DD1082 pKa = 3.55KK1083 pKa = 10.97TT1084 pKa = 3.75
Molecular weight: 125.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1906 |
410 |
1084 |
635.3 |
73.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.092 ± 0.101 | 1.574 ± 0.108 |
5.456 ± 0.52 | 6.139 ± 0.098 |
3.83 ± 0.181 | 3.253 ± 0.486 |
2.833 ± 0.311 | 7.975 ± 1.313 |
6.716 ± 0.882 | 9.286 ± 0.988 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.941 ± 0.22 | 5.981 ± 0.875 |
6.558 ± 1.684 | 5.247 ± 0.388 |
5.247 ± 0.74 | 6.716 ± 0.208 |
6.821 ± 0.165 | 5.299 ± 0.347 |
0.577 ± 0.098 | 4.46 ± 0.478 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
