
Rhizobium sp. AAP43
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
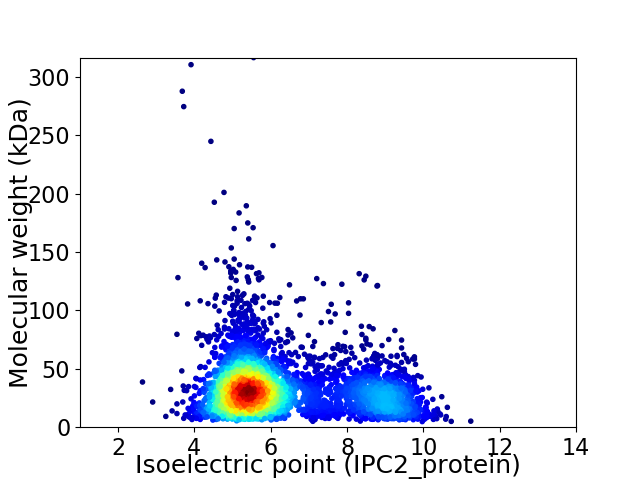
Virtual 2D-PAGE plot for 4372 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N0J4Y9|A0A0N0J4Y9_9RHIZ Ureidoacrylate amidohydrolase RutB OS=Rhizobium sp. AAP43 OX=1523420 GN=rutB PE=3 SV=1
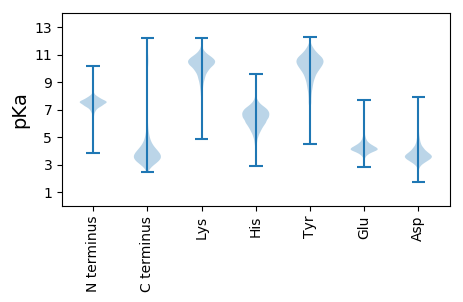
MM1 pKa = 7.38ARR3 pKa = 11.84LYY5 pKa = 10.24IEE7 pKa = 4.49SKK9 pKa = 10.4AVQQMTVWDD18 pKa = 4.01HH19 pKa = 6.55LYY21 pKa = 10.11IVYY24 pKa = 10.38EE25 pKa = 4.23DD26 pKa = 4.7DD27 pKa = 3.83FGQEE31 pKa = 3.95FVLRR35 pKa = 11.84AGPTSDD41 pKa = 5.65SPLDD45 pKa = 3.42WGSMEE50 pKa = 4.24VEE52 pKa = 4.09IGVPMALSEE61 pKa = 4.13DD62 pKa = 3.38ARR64 pKa = 11.84PIEE67 pKa = 4.84DD68 pKa = 3.62RR69 pKa = 11.84ALHH72 pKa = 6.29GNALLDD78 pKa = 3.79LAGRR82 pKa = 11.84DD83 pKa = 3.48ATAVFQTMMHH93 pKa = 6.38HH94 pKa = 7.35AGNIAQSLLPYY105 pKa = 10.28SAATQNSNSFVASLLHH121 pKa = 6.19VVGIDD126 pKa = 3.64VANVAPDD133 pKa = 3.23TGGIDD138 pKa = 3.31WTPASGNILDD148 pKa = 4.41SIEE151 pKa = 4.19FNVVGGYY158 pKa = 10.8DD159 pKa = 3.45DD160 pKa = 6.22FEE162 pKa = 4.83DD163 pKa = 4.22HH164 pKa = 7.53GDD166 pKa = 3.51ILRR169 pKa = 11.84GGALNDD175 pKa = 4.79AISGRR180 pKa = 11.84AGIDD184 pKa = 3.29QIDD187 pKa = 4.2GGDD190 pKa = 3.94GDD192 pKa = 4.2DD193 pKa = 3.61TLYY196 pKa = 10.87SGSVEE201 pKa = 4.45TPDD204 pKa = 4.63SDD206 pKa = 4.92ADD208 pKa = 3.85YY209 pKa = 9.0LTGGGGVDD217 pKa = 3.32NFYY220 pKa = 11.25VGNSDD225 pKa = 3.84YY226 pKa = 10.22EE227 pKa = 4.45TGLFRR232 pKa = 11.84YY233 pKa = 9.79DD234 pKa = 2.85VDD236 pKa = 3.76SGVMYY241 pKa = 10.46FNSEE245 pKa = 3.51SRR247 pKa = 11.84GRR249 pKa = 11.84YY250 pKa = 9.0DD251 pKa = 3.49VIVDD255 pKa = 4.38FEE257 pKa = 4.59SSDD260 pKa = 4.03AIHH263 pKa = 6.48VSGFGVWDD271 pKa = 3.49YY272 pKa = 11.76TEE274 pKa = 4.87FSSITIDD281 pKa = 3.47DD282 pKa = 3.63AGYY285 pKa = 10.62AIDD288 pKa = 4.78GKK290 pKa = 10.36DD291 pKa = 3.17VYY293 pKa = 10.52VANGSGLEE301 pKa = 4.03LFAVRR306 pKa = 11.84DD307 pKa = 3.71SYY309 pKa = 11.93FDD311 pKa = 3.53VNLEE315 pKa = 3.73RR316 pKa = 11.84DD317 pKa = 3.21IDD319 pKa = 4.56VMVFWYY325 pKa = 10.26QISSRR330 pKa = 11.84MSGMDD335 pKa = 3.72DD336 pKa = 3.33VLIDD340 pKa = 3.59VNIPLFAIDD349 pKa = 4.98YY350 pKa = 7.6GSPWFQQSSSSSFSTADD367 pKa = 3.03ASGSSDD373 pKa = 3.5SIVFQPEE380 pKa = 4.05LVAEE384 pKa = 4.58APSNGYY390 pKa = 9.08DD391 pKa = 3.1QSDD394 pKa = 3.54FSEE397 pKa = 4.75LDD399 pKa = 3.31GLSEE403 pKa = 4.56FGVSHH408 pKa = 6.23ITDD411 pKa = 3.94SIYY414 pKa = 10.92FEE416 pKa = 5.77DD417 pKa = 4.79GFSGIQVHH425 pKa = 7.36DD426 pKa = 4.19GLEE429 pKa = 4.32STHH432 pKa = 5.97QRR434 pKa = 11.84LDD436 pKa = 3.09VDD438 pKa = 3.81EE439 pKa = 4.96FRR441 pKa = 11.84LVAA444 pKa = 4.74
MM1 pKa = 7.38ARR3 pKa = 11.84LYY5 pKa = 10.24IEE7 pKa = 4.49SKK9 pKa = 10.4AVQQMTVWDD18 pKa = 4.01HH19 pKa = 6.55LYY21 pKa = 10.11IVYY24 pKa = 10.38EE25 pKa = 4.23DD26 pKa = 4.7DD27 pKa = 3.83FGQEE31 pKa = 3.95FVLRR35 pKa = 11.84AGPTSDD41 pKa = 5.65SPLDD45 pKa = 3.42WGSMEE50 pKa = 4.24VEE52 pKa = 4.09IGVPMALSEE61 pKa = 4.13DD62 pKa = 3.38ARR64 pKa = 11.84PIEE67 pKa = 4.84DD68 pKa = 3.62RR69 pKa = 11.84ALHH72 pKa = 6.29GNALLDD78 pKa = 3.79LAGRR82 pKa = 11.84DD83 pKa = 3.48ATAVFQTMMHH93 pKa = 6.38HH94 pKa = 7.35AGNIAQSLLPYY105 pKa = 10.28SAATQNSNSFVASLLHH121 pKa = 6.19VVGIDD126 pKa = 3.64VANVAPDD133 pKa = 3.23TGGIDD138 pKa = 3.31WTPASGNILDD148 pKa = 4.41SIEE151 pKa = 4.19FNVVGGYY158 pKa = 10.8DD159 pKa = 3.45DD160 pKa = 6.22FEE162 pKa = 4.83DD163 pKa = 4.22HH164 pKa = 7.53GDD166 pKa = 3.51ILRR169 pKa = 11.84GGALNDD175 pKa = 4.79AISGRR180 pKa = 11.84AGIDD184 pKa = 3.29QIDD187 pKa = 4.2GGDD190 pKa = 3.94GDD192 pKa = 4.2DD193 pKa = 3.61TLYY196 pKa = 10.87SGSVEE201 pKa = 4.45TPDD204 pKa = 4.63SDD206 pKa = 4.92ADD208 pKa = 3.85YY209 pKa = 9.0LTGGGGVDD217 pKa = 3.32NFYY220 pKa = 11.25VGNSDD225 pKa = 3.84YY226 pKa = 10.22EE227 pKa = 4.45TGLFRR232 pKa = 11.84YY233 pKa = 9.79DD234 pKa = 2.85VDD236 pKa = 3.76SGVMYY241 pKa = 10.46FNSEE245 pKa = 3.51SRR247 pKa = 11.84GRR249 pKa = 11.84YY250 pKa = 9.0DD251 pKa = 3.49VIVDD255 pKa = 4.38FEE257 pKa = 4.59SSDD260 pKa = 4.03AIHH263 pKa = 6.48VSGFGVWDD271 pKa = 3.49YY272 pKa = 11.76TEE274 pKa = 4.87FSSITIDD281 pKa = 3.47DD282 pKa = 3.63AGYY285 pKa = 10.62AIDD288 pKa = 4.78GKK290 pKa = 10.36DD291 pKa = 3.17VYY293 pKa = 10.52VANGSGLEE301 pKa = 4.03LFAVRR306 pKa = 11.84DD307 pKa = 3.71SYY309 pKa = 11.93FDD311 pKa = 3.53VNLEE315 pKa = 3.73RR316 pKa = 11.84DD317 pKa = 3.21IDD319 pKa = 4.56VMVFWYY325 pKa = 10.26QISSRR330 pKa = 11.84MSGMDD335 pKa = 3.72DD336 pKa = 3.33VLIDD340 pKa = 3.59VNIPLFAIDD349 pKa = 4.98YY350 pKa = 7.6GSPWFQQSSSSSFSTADD367 pKa = 3.03ASGSSDD373 pKa = 3.5SIVFQPEE380 pKa = 4.05LVAEE384 pKa = 4.58APSNGYY390 pKa = 9.08DD391 pKa = 3.1QSDD394 pKa = 3.54FSEE397 pKa = 4.75LDD399 pKa = 3.31GLSEE403 pKa = 4.56FGVSHH408 pKa = 6.23ITDD411 pKa = 3.94SIYY414 pKa = 10.92FEE416 pKa = 5.77DD417 pKa = 4.79GFSGIQVHH425 pKa = 7.36DD426 pKa = 4.19GLEE429 pKa = 4.32STHH432 pKa = 5.97QRR434 pKa = 11.84LDD436 pKa = 3.09VDD438 pKa = 3.81EE439 pKa = 4.96FRR441 pKa = 11.84LVAA444 pKa = 4.74
Molecular weight: 48.21 kDa
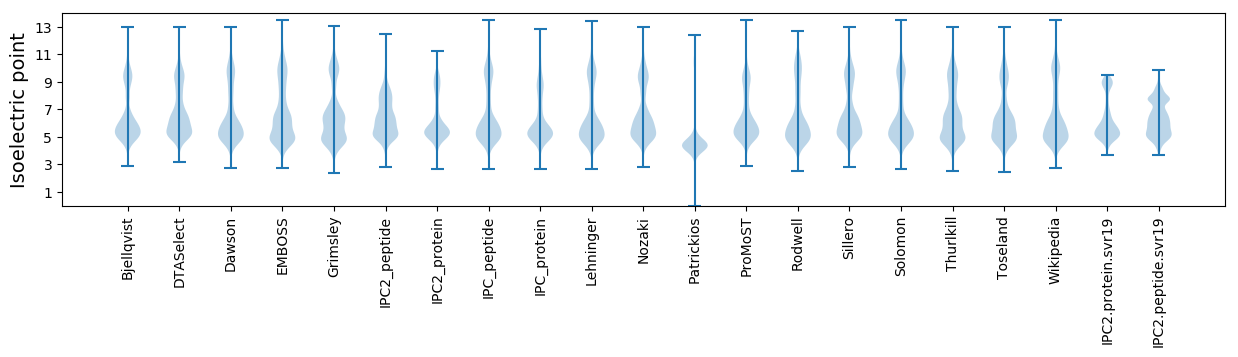
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N0JB90|A0A0N0JB90_9RHIZ Calcium-binding protein OS=Rhizobium sp. AAP43 OX=1523420 GN=IP76_00665 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.84LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.43GGRR29 pKa = 11.84QVLNARR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84GRR40 pKa = 11.84AKK42 pKa = 10.69LSAA45 pKa = 4.0
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.84LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.43GGRR29 pKa = 11.84QVLNARR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84GRR40 pKa = 11.84AKK42 pKa = 10.69LSAA45 pKa = 4.0
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394556 |
41 |
2949 |
319.0 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.882 ± 0.048 | 0.787 ± 0.013 |
5.835 ± 0.032 | 5.86 ± 0.034 |
3.885 ± 0.022 | 8.34 ± 0.036 |
2.003 ± 0.018 | 5.635 ± 0.03 |
3.578 ± 0.029 | 10.116 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.019 | 2.734 ± 0.024 |
4.814 ± 0.031 | 3.173 ± 0.023 |
6.575 ± 0.044 | 5.793 ± 0.031 |
5.482 ± 0.031 | 7.365 ± 0.03 |
1.243 ± 0.015 | 2.269 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
