
Vibrio phage K05K4_VK05K4_2
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
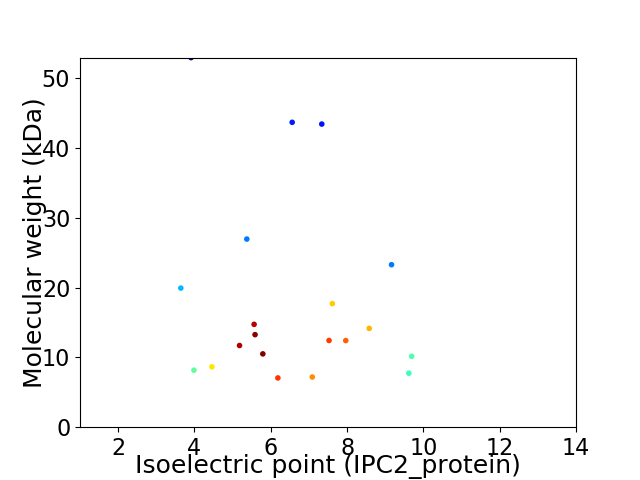
Virtual 2D-PAGE plot for 20 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6UG32|A0A1W6UG32_9VIRU Minor capsid protein OS=Vibrio phage K05K4_VK05K4_2 OX=1912327 GN=K05K4_52820 PE=4 SV=1
MM1 pKa = 7.29FLDD4 pKa = 3.39TGYY7 pKa = 11.16FDD9 pKa = 3.34TCTFEE14 pKa = 3.83KK15 pKa = 10.29TPYY18 pKa = 10.41SNARR22 pKa = 11.84YY23 pKa = 8.73PYY25 pKa = 9.25QTVCDD30 pKa = 3.76NGLGLGYY37 pKa = 10.63YY38 pKa = 7.75EE39 pKa = 4.43VRR41 pKa = 11.84CPEE44 pKa = 3.81NSEE47 pKa = 4.27FDD49 pKa = 3.99PSTLRR54 pKa = 11.84CKK56 pKa = 10.33SVCEE60 pKa = 4.04YY61 pKa = 11.13GKK63 pKa = 10.26NPDD66 pKa = 3.94GTCMDD71 pKa = 3.88ACQFKK76 pKa = 10.92KK77 pKa = 10.76SIDD80 pKa = 4.02EE81 pKa = 4.15IKK83 pKa = 10.48SLQWLAYY90 pKa = 9.88VYY92 pKa = 10.64GEE94 pKa = 4.11QVTGSCYY101 pKa = 10.68GDD103 pKa = 3.56YY104 pKa = 11.09GATRR108 pKa = 11.84CEE110 pKa = 4.25LEE112 pKa = 4.18RR113 pKa = 11.84TPSDD117 pKa = 3.21STLCTGVDD125 pKa = 3.31SGQWTQNTICHH136 pKa = 6.32GNFQFTGNQCEE147 pKa = 4.28GGTLFWGKK155 pKa = 10.11DD156 pKa = 3.62GPDD159 pKa = 3.31TPIIPDD165 pKa = 4.37DD166 pKa = 5.14PIHH169 pKa = 7.61DD170 pKa = 4.88PDD172 pKa = 5.88DD173 pKa = 3.89PTGDD177 pKa = 3.64IEE179 pKa = 5.54DD180 pKa = 4.5PSILPDD186 pKa = 3.61GSTNTVNPPDD196 pKa = 4.02TDD198 pKa = 3.65SEE200 pKa = 4.63PDD202 pKa = 3.48VEE204 pKa = 5.55EE205 pKa = 5.22PDD207 pKa = 3.63TDD209 pKa = 3.49EE210 pKa = 4.45STDD213 pKa = 3.64TAVLKK218 pKa = 10.91AITGMNKK225 pKa = 10.0DD226 pKa = 3.38VNKK229 pKa = 10.54ALNDD233 pKa = 3.47MNIDD237 pKa = 3.4INQANADD244 pKa = 3.75VQNQIIALNASMVTNTQAIQKK265 pKa = 7.84QQINDD270 pKa = 3.14NKK272 pKa = 10.37IYY274 pKa = 10.49EE275 pKa = 4.16NTKK278 pKa = 10.95ALIQQANADD287 pKa = 3.49ITTAMNKK294 pKa = 7.52NTNAVNGVGDD304 pKa = 4.45DD305 pKa = 3.72VEE307 pKa = 5.55KK308 pKa = 10.74IAGAMDD314 pKa = 5.45GIAEE318 pKa = 4.26DD319 pKa = 3.65VSGISDD325 pKa = 3.9TLDD328 pKa = 4.1GIANTDD334 pKa = 3.08TSGAGTGGTCIEE346 pKa = 4.43SQSCTGFYY354 pKa = 10.69EE355 pKa = 4.04SGYY358 pKa = 9.68PDD360 pKa = 3.62GLGGLVSGQLDD371 pKa = 3.73DD372 pKa = 5.58LKK374 pKa = 11.35HH375 pKa = 4.84NTIDD379 pKa = 3.42NFVNSFGDD387 pKa = 3.73LDD389 pKa = 4.02LSSAKK394 pKa = 10.1RR395 pKa = 11.84PSFVLPVPFFGDD407 pKa = 3.76FSFEE411 pKa = 4.02EE412 pKa = 4.39QISFDD417 pKa = 3.32WVFGFIRR424 pKa = 11.84AVLIMTSVFAARR436 pKa = 11.84RR437 pKa = 11.84IILEE441 pKa = 4.04VNMDD445 pKa = 3.55WLVDD449 pKa = 4.05LFNKK453 pKa = 10.38LLVFLYY459 pKa = 10.34QLLISLVNMLKK470 pKa = 10.66DD471 pKa = 3.49LFFGRR476 pKa = 11.84LSKK479 pKa = 10.26SWQWW483 pKa = 2.99
MM1 pKa = 7.29FLDD4 pKa = 3.39TGYY7 pKa = 11.16FDD9 pKa = 3.34TCTFEE14 pKa = 3.83KK15 pKa = 10.29TPYY18 pKa = 10.41SNARR22 pKa = 11.84YY23 pKa = 8.73PYY25 pKa = 9.25QTVCDD30 pKa = 3.76NGLGLGYY37 pKa = 10.63YY38 pKa = 7.75EE39 pKa = 4.43VRR41 pKa = 11.84CPEE44 pKa = 3.81NSEE47 pKa = 4.27FDD49 pKa = 3.99PSTLRR54 pKa = 11.84CKK56 pKa = 10.33SVCEE60 pKa = 4.04YY61 pKa = 11.13GKK63 pKa = 10.26NPDD66 pKa = 3.94GTCMDD71 pKa = 3.88ACQFKK76 pKa = 10.92KK77 pKa = 10.76SIDD80 pKa = 4.02EE81 pKa = 4.15IKK83 pKa = 10.48SLQWLAYY90 pKa = 9.88VYY92 pKa = 10.64GEE94 pKa = 4.11QVTGSCYY101 pKa = 10.68GDD103 pKa = 3.56YY104 pKa = 11.09GATRR108 pKa = 11.84CEE110 pKa = 4.25LEE112 pKa = 4.18RR113 pKa = 11.84TPSDD117 pKa = 3.21STLCTGVDD125 pKa = 3.31SGQWTQNTICHH136 pKa = 6.32GNFQFTGNQCEE147 pKa = 4.28GGTLFWGKK155 pKa = 10.11DD156 pKa = 3.62GPDD159 pKa = 3.31TPIIPDD165 pKa = 4.37DD166 pKa = 5.14PIHH169 pKa = 7.61DD170 pKa = 4.88PDD172 pKa = 5.88DD173 pKa = 3.89PTGDD177 pKa = 3.64IEE179 pKa = 5.54DD180 pKa = 4.5PSILPDD186 pKa = 3.61GSTNTVNPPDD196 pKa = 4.02TDD198 pKa = 3.65SEE200 pKa = 4.63PDD202 pKa = 3.48VEE204 pKa = 5.55EE205 pKa = 5.22PDD207 pKa = 3.63TDD209 pKa = 3.49EE210 pKa = 4.45STDD213 pKa = 3.64TAVLKK218 pKa = 10.91AITGMNKK225 pKa = 10.0DD226 pKa = 3.38VNKK229 pKa = 10.54ALNDD233 pKa = 3.47MNIDD237 pKa = 3.4INQANADD244 pKa = 3.75VQNQIIALNASMVTNTQAIQKK265 pKa = 7.84QQINDD270 pKa = 3.14NKK272 pKa = 10.37IYY274 pKa = 10.49EE275 pKa = 4.16NTKK278 pKa = 10.95ALIQQANADD287 pKa = 3.49ITTAMNKK294 pKa = 7.52NTNAVNGVGDD304 pKa = 4.45DD305 pKa = 3.72VEE307 pKa = 5.55KK308 pKa = 10.74IAGAMDD314 pKa = 5.45GIAEE318 pKa = 4.26DD319 pKa = 3.65VSGISDD325 pKa = 3.9TLDD328 pKa = 4.1GIANTDD334 pKa = 3.08TSGAGTGGTCIEE346 pKa = 4.43SQSCTGFYY354 pKa = 10.69EE355 pKa = 4.04SGYY358 pKa = 9.68PDD360 pKa = 3.62GLGGLVSGQLDD371 pKa = 3.73DD372 pKa = 5.58LKK374 pKa = 11.35HH375 pKa = 4.84NTIDD379 pKa = 3.42NFVNSFGDD387 pKa = 3.73LDD389 pKa = 4.02LSSAKK394 pKa = 10.1RR395 pKa = 11.84PSFVLPVPFFGDD407 pKa = 3.76FSFEE411 pKa = 4.02EE412 pKa = 4.39QISFDD417 pKa = 3.32WVFGFIRR424 pKa = 11.84AVLIMTSVFAARR436 pKa = 11.84RR437 pKa = 11.84IILEE441 pKa = 4.04VNMDD445 pKa = 3.55WLVDD449 pKa = 4.05LFNKK453 pKa = 10.38LLVFLYY459 pKa = 10.34QLLISLVNMLKK470 pKa = 10.66DD471 pKa = 3.49LFFGRR476 pKa = 11.84LSKK479 pKa = 10.26SWQWW483 pKa = 2.99
Molecular weight: 52.99 kDa
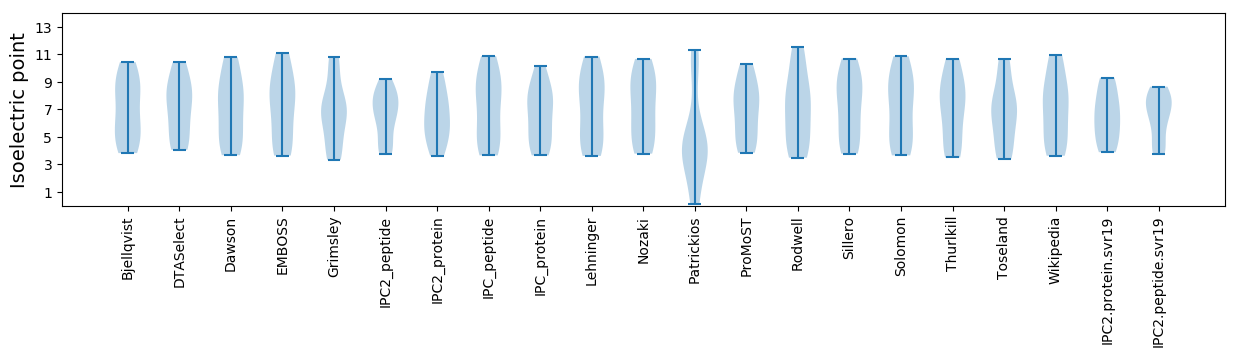
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6UH42|A0A1W6UH42_9VIRU Uncharacterized protein OS=Vibrio phage K05K4_VK05K4_2 OX=1912327 GN=K05K4_53010 PE=4 SV=1
MM1 pKa = 6.84VRR3 pKa = 11.84SVSRR7 pKa = 11.84SMPTPTKK14 pKa = 9.78WIRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 9.55LKK21 pKa = 9.52KK22 pKa = 10.75LGVRR26 pKa = 11.84RR27 pKa = 11.84VGKK30 pKa = 10.09SLSNGVLFTGDD41 pKa = 3.51AVTEE45 pKa = 4.07ILTCLQAKK53 pKa = 7.22QAKK56 pKa = 9.55NDD58 pKa = 4.19TIPFEE63 pKa = 4.71LNISALPILTWFRR76 pKa = 11.84SPLSLGTPTLEE87 pKa = 4.14AQYY90 pKa = 11.37SS91 pKa = 3.66
MM1 pKa = 6.84VRR3 pKa = 11.84SVSRR7 pKa = 11.84SMPTPTKK14 pKa = 9.78WIRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 9.55LKK21 pKa = 9.52KK22 pKa = 10.75LGVRR26 pKa = 11.84RR27 pKa = 11.84VGKK30 pKa = 10.09SLSNGVLFTGDD41 pKa = 3.51AVTEE45 pKa = 4.07ILTCLQAKK53 pKa = 7.22QAKK56 pKa = 9.55NDD58 pKa = 4.19TIPFEE63 pKa = 4.71LNISALPILTWFRR76 pKa = 11.84SPLSLGTPTLEE87 pKa = 4.14AQYY90 pKa = 11.37SS91 pKa = 3.66
Molecular weight: 10.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3264 |
61 |
483 |
163.2 |
18.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.893 ± 0.618 | 2.145 ± 0.392 |
6.434 ± 0.766 | 5.392 ± 0.314 |
4.32 ± 0.281 | 6.648 ± 0.448 |
1.961 ± 0.345 | 6.036 ± 0.38 |
6.005 ± 0.614 | 9.283 ± 0.735 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.911 ± 0.44 | 4.688 ± 0.604 |
4.044 ± 0.345 | 4.32 ± 0.561 |
4.381 ± 0.57 | 6.71 ± 0.464 |
6.311 ± 0.593 | 6.311 ± 0.384 |
1.409 ± 0.186 | 3.799 ± 0.412 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
