
Tortoise microvirus 40
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
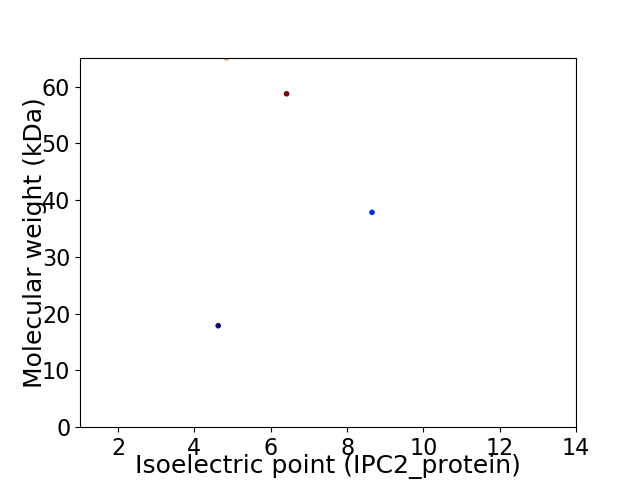
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
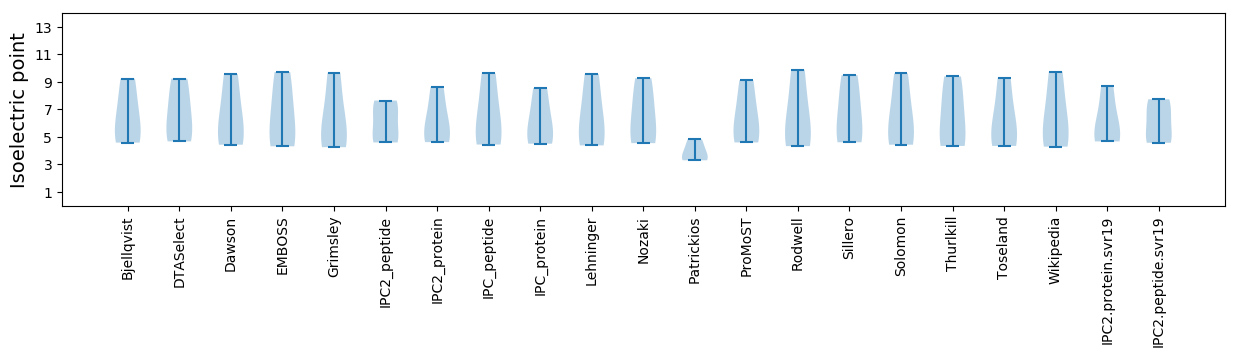
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6A4|A0A4P8W6A4_9VIRU Major capsid protein OS=Tortoise microvirus 40 OX=2583144 PE=3 SV=1
MM1 pKa = 7.12FSKK4 pKa = 10.54RR5 pKa = 11.84RR6 pKa = 11.84TTPYY10 pKa = 10.54VYY12 pKa = 10.78VDD14 pKa = 3.93DD15 pKa = 4.4PVIVSSKK22 pKa = 10.88EE23 pKa = 3.93MICSEE28 pKa = 4.44FKK30 pKa = 11.03EE31 pKa = 4.33EE32 pKa = 4.23TPVDD36 pKa = 3.39QFLQQEE42 pKa = 4.29IEE44 pKa = 4.37VNGVKK49 pKa = 10.34SIRR52 pKa = 11.84LTSDD56 pKa = 1.95IYY58 pKa = 11.07MLFNQQRR65 pKa = 11.84LDD67 pKa = 3.49RR68 pKa = 11.84LSRR71 pKa = 11.84EE72 pKa = 3.74SLLAYY77 pKa = 10.34FEE79 pKa = 4.99SISVNEE85 pKa = 4.29PKK87 pKa = 10.74FGDD90 pKa = 3.72LRR92 pKa = 11.84SKK94 pKa = 11.05LGDD97 pKa = 3.57DD98 pKa = 3.51QLISFVKK105 pKa = 10.43SRR107 pKa = 11.84FIQSPSEE114 pKa = 3.7LMAWSSYY121 pKa = 10.62LMSLMSSSDD130 pKa = 3.15AAIAEE135 pKa = 4.32LSALQSQVQPEE146 pKa = 4.52AQPEE150 pKa = 4.17AQPEE154 pKa = 4.12AQPP157 pKa = 3.74
MM1 pKa = 7.12FSKK4 pKa = 10.54RR5 pKa = 11.84RR6 pKa = 11.84TTPYY10 pKa = 10.54VYY12 pKa = 10.78VDD14 pKa = 3.93DD15 pKa = 4.4PVIVSSKK22 pKa = 10.88EE23 pKa = 3.93MICSEE28 pKa = 4.44FKK30 pKa = 11.03EE31 pKa = 4.33EE32 pKa = 4.23TPVDD36 pKa = 3.39QFLQQEE42 pKa = 4.29IEE44 pKa = 4.37VNGVKK49 pKa = 10.34SIRR52 pKa = 11.84LTSDD56 pKa = 1.95IYY58 pKa = 11.07MLFNQQRR65 pKa = 11.84LDD67 pKa = 3.49RR68 pKa = 11.84LSRR71 pKa = 11.84EE72 pKa = 3.74SLLAYY77 pKa = 10.34FEE79 pKa = 4.99SISVNEE85 pKa = 4.29PKK87 pKa = 10.74FGDD90 pKa = 3.72LRR92 pKa = 11.84SKK94 pKa = 11.05LGDD97 pKa = 3.57DD98 pKa = 3.51QLISFVKK105 pKa = 10.43SRR107 pKa = 11.84FIQSPSEE114 pKa = 3.7LMAWSSYY121 pKa = 10.62LMSLMSSSDD130 pKa = 3.15AAIAEE135 pKa = 4.32LSALQSQVQPEE146 pKa = 4.52AQPEE150 pKa = 4.17AQPEE154 pKa = 4.12AQPP157 pKa = 3.74
Molecular weight: 17.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W641|A0A4P8W641_9VIRU Internal scaffolding protein OS=Tortoise microvirus 40 OX=2583144 PE=4 SV=1
MM1 pKa = 7.87AIGATAASLIAAGVGAVGSTAGAVLSNKK29 pKa = 8.18GTASIANNNNNFSEE43 pKa = 4.28KK44 pKa = 10.21MLDD47 pKa = 3.44KK48 pKa = 10.9QIAYY52 pKa = 9.63NKK54 pKa = 10.07EE55 pKa = 3.96MYY57 pKa = 9.3QQQLGDD63 pKa = 3.04QWQFYY68 pKa = 11.14NDD70 pKa = 3.43AKK72 pKa = 10.52QNAWDD77 pKa = 3.59MAEE80 pKa = 4.29YY81 pKa = 10.8NSAPAQRR88 pKa = 11.84EE89 pKa = 3.96RR90 pKa = 11.84LEE92 pKa = 4.2AAGLNPYY99 pKa = 11.24LMMNGGSVGTAQSSAQGSAPSAQGVNPPTATPYY132 pKa = 11.35SMDD135 pKa = 3.13YY136 pKa = 10.75SGLVQGLGMALDD148 pKa = 4.86QISKK152 pKa = 9.58MPDD155 pKa = 2.91NSVKK159 pKa = 10.5RR160 pKa = 11.84AEE162 pKa = 4.27ADD164 pKa = 3.14NLRR167 pKa = 11.84IEE169 pKa = 4.8GKK171 pKa = 9.98YY172 pKa = 9.72KK173 pKa = 9.12AAKK176 pKa = 8.41MVAEE180 pKa = 4.63IAQMRR185 pKa = 11.84TNAHH189 pKa = 4.54TQKK192 pKa = 10.7GRR194 pKa = 11.84LALDD198 pKa = 3.27KK199 pKa = 10.8LIYY202 pKa = 10.82SIDD205 pKa = 3.82KK206 pKa = 10.4DD207 pKa = 3.86LKK209 pKa = 8.11TSQMSVNSEE218 pKa = 4.28NIANMQAQRR227 pKa = 11.84KK228 pKa = 8.28LINVQTLFAEE238 pKa = 4.78KK239 pKa = 10.05QLSWMDD245 pKa = 3.56AQNKK249 pKa = 8.51MDD251 pKa = 5.14LAQKK255 pKa = 8.87TADD258 pKa = 3.4IQLKK262 pKa = 8.49YY263 pKa = 10.67AQGALTKK270 pKa = 10.2KK271 pKa = 9.88QVEE274 pKa = 4.29HH275 pKa = 6.4EE276 pKa = 4.37VNKK279 pKa = 10.26IAEE282 pKa = 4.27TAARR286 pKa = 11.84TSLIGVQKK294 pKa = 10.49QGQSLQNQFDD304 pKa = 3.79SATFGNRR311 pKa = 11.84VKK313 pKa = 9.82TVKK316 pKa = 9.55ATLYY320 pKa = 10.92NLISDD325 pKa = 4.79AYY327 pKa = 9.6SLPKK331 pKa = 10.35AAGSYY336 pKa = 10.38FMNTHH341 pKa = 4.83IQKK344 pKa = 10.2EE345 pKa = 4.3FFKK348 pKa = 11.15
MM1 pKa = 7.87AIGATAASLIAAGVGAVGSTAGAVLSNKK29 pKa = 8.18GTASIANNNNNFSEE43 pKa = 4.28KK44 pKa = 10.21MLDD47 pKa = 3.44KK48 pKa = 10.9QIAYY52 pKa = 9.63NKK54 pKa = 10.07EE55 pKa = 3.96MYY57 pKa = 9.3QQQLGDD63 pKa = 3.04QWQFYY68 pKa = 11.14NDD70 pKa = 3.43AKK72 pKa = 10.52QNAWDD77 pKa = 3.59MAEE80 pKa = 4.29YY81 pKa = 10.8NSAPAQRR88 pKa = 11.84EE89 pKa = 3.96RR90 pKa = 11.84LEE92 pKa = 4.2AAGLNPYY99 pKa = 11.24LMMNGGSVGTAQSSAQGSAPSAQGVNPPTATPYY132 pKa = 11.35SMDD135 pKa = 3.13YY136 pKa = 10.75SGLVQGLGMALDD148 pKa = 4.86QISKK152 pKa = 9.58MPDD155 pKa = 2.91NSVKK159 pKa = 10.5RR160 pKa = 11.84AEE162 pKa = 4.27ADD164 pKa = 3.14NLRR167 pKa = 11.84IEE169 pKa = 4.8GKK171 pKa = 9.98YY172 pKa = 9.72KK173 pKa = 9.12AAKK176 pKa = 8.41MVAEE180 pKa = 4.63IAQMRR185 pKa = 11.84TNAHH189 pKa = 4.54TQKK192 pKa = 10.7GRR194 pKa = 11.84LALDD198 pKa = 3.27KK199 pKa = 10.8LIYY202 pKa = 10.82SIDD205 pKa = 3.82KK206 pKa = 10.4DD207 pKa = 3.86LKK209 pKa = 8.11TSQMSVNSEE218 pKa = 4.28NIANMQAQRR227 pKa = 11.84KK228 pKa = 8.28LINVQTLFAEE238 pKa = 4.78KK239 pKa = 10.05QLSWMDD245 pKa = 3.56AQNKK249 pKa = 8.51MDD251 pKa = 5.14LAQKK255 pKa = 8.87TADD258 pKa = 3.4IQLKK262 pKa = 8.49YY263 pKa = 10.67AQGALTKK270 pKa = 10.2KK271 pKa = 9.88QVEE274 pKa = 4.29HH275 pKa = 6.4EE276 pKa = 4.37VNKK279 pKa = 10.26IAEE282 pKa = 4.27TAARR286 pKa = 11.84TSLIGVQKK294 pKa = 10.49QGQSLQNQFDD304 pKa = 3.79SATFGNRR311 pKa = 11.84VKK313 pKa = 9.82TVKK316 pKa = 9.55ATLYY320 pKa = 10.92NLISDD325 pKa = 4.79AYY327 pKa = 9.6SLPKK331 pKa = 10.35AAGSYY336 pKa = 10.38FMNTHH341 pKa = 4.83IQKK344 pKa = 10.2EE345 pKa = 4.3FFKK348 pKa = 11.15
Molecular weight: 37.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1582 |
157 |
580 |
395.5 |
44.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.143 ± 1.916 | 1.517 ± 0.724 |
6.384 ± 0.556 | 4.551 ± 0.982 |
6.131 ± 1.184 | 4.867 ± 0.882 |
1.58 ± 0.605 | 4.298 ± 0.278 |
5.942 ± 0.788 | 8.534 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.592 ± 0.694 | 6.384 ± 1.369 |
3.666 ± 0.441 | 5.563 ± 1.168 |
3.982 ± 0.629 | 9.735 ± 0.931 |
4.741 ± 0.371 | 5.879 ± 0.465 |
0.948 ± 0.114 | 5.563 ± 0.866 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
