
Cyprinid herpesvirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Cyprinivirus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
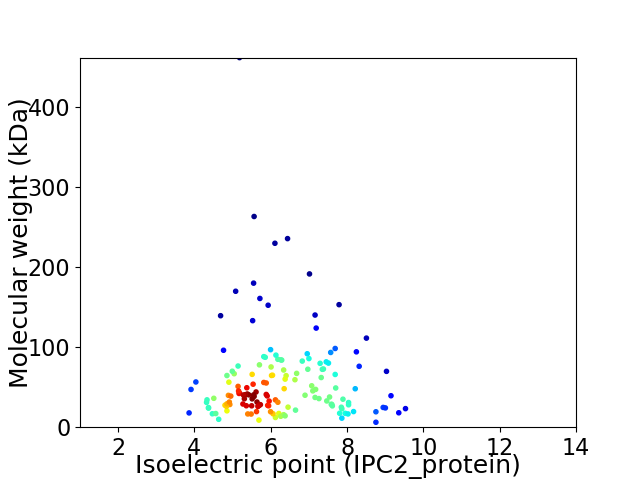
Virtual 2D-PAGE plot for 150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7PC21|K7PC21_9VIRU Protein ORF156 OS=Cyprinid herpesvirus 2 OX=317878 GN=CyHV2_ORF156 PE=4 SV=1
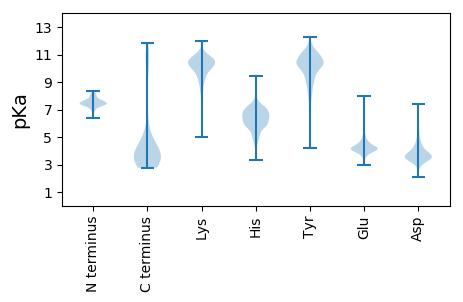
MM1 pKa = 7.39ARR3 pKa = 11.84CDD5 pKa = 5.18FFKK8 pKa = 10.61QWLLSSLPFILCIIGEE24 pKa = 4.3YY25 pKa = 10.53LVVGALAVAIIISSGIMVKK44 pKa = 10.55DD45 pKa = 3.86IDD47 pKa = 4.05DD48 pKa = 3.64QQATGEE54 pKa = 4.25CWPAVLLSGSGLFLTRR70 pKa = 11.84FWTVHH75 pKa = 5.38NYY77 pKa = 8.9QFGRR81 pKa = 11.84TRR83 pKa = 11.84TILGTRR89 pKa = 11.84SRR91 pKa = 11.84RR92 pKa = 11.84MRR94 pKa = 11.84FAAVILAIFVVASGVMSIWASIGDD118 pKa = 3.63MVMTYY123 pKa = 10.08MNDD126 pKa = 3.23HH127 pKa = 7.11ANPKK131 pKa = 8.23PTLDD135 pKa = 3.75QLEE138 pKa = 4.24QQEE141 pKa = 4.49KK142 pKa = 10.59DD143 pKa = 3.78VPCLFRR149 pKa = 11.84CYY151 pKa = 10.43GGYY154 pKa = 8.06HH155 pKa = 5.63TSIVLYY161 pKa = 10.19TMLAAGFCLLAIGLEE176 pKa = 4.22MEE178 pKa = 4.8DD179 pKa = 3.28RR180 pKa = 11.84AVDD183 pKa = 3.63AEE185 pKa = 4.13EE186 pKa = 4.5DD187 pKa = 3.66ALIGSTASAFTPDD200 pKa = 3.15STVDD204 pKa = 3.41LTAIPPPLPPPYY216 pKa = 10.14NGDD219 pKa = 3.58GVPPPSYY226 pKa = 10.72TNIINIYY233 pKa = 8.33TGNNGSGSEE242 pKa = 4.34APPPSYY248 pKa = 10.29EE249 pKa = 4.01VAVGEE254 pKa = 4.31TSQPEE259 pKa = 4.34EE260 pKa = 4.06PQQDD264 pKa = 3.47SSEE267 pKa = 4.12PQPDD271 pKa = 3.27SSEE274 pKa = 4.15PQPEE278 pKa = 3.83TRR280 pKa = 11.84IIITMM285 pKa = 4.39
MM1 pKa = 7.39ARR3 pKa = 11.84CDD5 pKa = 5.18FFKK8 pKa = 10.61QWLLSSLPFILCIIGEE24 pKa = 4.3YY25 pKa = 10.53LVVGALAVAIIISSGIMVKK44 pKa = 10.55DD45 pKa = 3.86IDD47 pKa = 4.05DD48 pKa = 3.64QQATGEE54 pKa = 4.25CWPAVLLSGSGLFLTRR70 pKa = 11.84FWTVHH75 pKa = 5.38NYY77 pKa = 8.9QFGRR81 pKa = 11.84TRR83 pKa = 11.84TILGTRR89 pKa = 11.84SRR91 pKa = 11.84RR92 pKa = 11.84MRR94 pKa = 11.84FAAVILAIFVVASGVMSIWASIGDD118 pKa = 3.63MVMTYY123 pKa = 10.08MNDD126 pKa = 3.23HH127 pKa = 7.11ANPKK131 pKa = 8.23PTLDD135 pKa = 3.75QLEE138 pKa = 4.24QQEE141 pKa = 4.49KK142 pKa = 10.59DD143 pKa = 3.78VPCLFRR149 pKa = 11.84CYY151 pKa = 10.43GGYY154 pKa = 8.06HH155 pKa = 5.63TSIVLYY161 pKa = 10.19TMLAAGFCLLAIGLEE176 pKa = 4.22MEE178 pKa = 4.8DD179 pKa = 3.28RR180 pKa = 11.84AVDD183 pKa = 3.63AEE185 pKa = 4.13EE186 pKa = 4.5DD187 pKa = 3.66ALIGSTASAFTPDD200 pKa = 3.15STVDD204 pKa = 3.41LTAIPPPLPPPYY216 pKa = 10.14NGDD219 pKa = 3.58GVPPPSYY226 pKa = 10.72TNIINIYY233 pKa = 8.33TGNNGSGSEE242 pKa = 4.34APPPSYY248 pKa = 10.29EE249 pKa = 4.01VAVGEE254 pKa = 4.31TSQPEE259 pKa = 4.34EE260 pKa = 4.06PQQDD264 pKa = 3.47SSEE267 pKa = 4.12PQPDD271 pKa = 3.27SSEE274 pKa = 4.15PQPEE278 pKa = 3.83TRR280 pKa = 11.84IIITMM285 pKa = 4.39
Molecular weight: 31.02 kDa
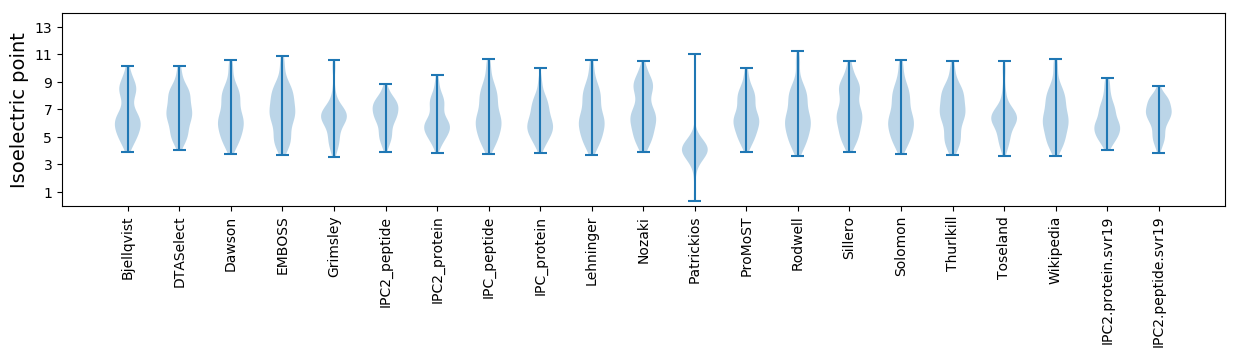
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7PBQ8|K7PBQ8_9VIRU ORF2A OS=Cyprinid herpesvirus 2 OX=317878 GN=CyHV2_149 PE=4 SV=1
MM1 pKa = 7.4VYY3 pKa = 10.67YY4 pKa = 10.08ILLLGLVGLGLGEE17 pKa = 4.3STNTSSSNSSSIASSLPPPKK37 pKa = 8.85ITHH40 pKa = 6.47IPTTTSPTVTVTVTDD55 pKa = 3.98LMLGLVVLLLVVILSLLVVILLTLKK80 pKa = 10.64RR81 pKa = 11.84MRR83 pKa = 11.84KK84 pKa = 8.85DD85 pKa = 3.09AKK87 pKa = 10.71AEE89 pKa = 3.82KK90 pKa = 10.08DD91 pKa = 3.52KK92 pKa = 11.15KK93 pKa = 10.55QKK95 pKa = 10.29PDD97 pKa = 3.69DD98 pKa = 3.86VEE100 pKa = 4.05MSLLRR105 pKa = 11.84RR106 pKa = 11.84MHH108 pKa = 6.77SDD110 pKa = 2.72HH111 pKa = 5.86MHH113 pKa = 5.21MHH115 pKa = 6.32SEE117 pKa = 3.82AAGPFGNMYY126 pKa = 8.94TRR128 pKa = 11.84PLPSPPQNQWWQNGTLGGRR147 pKa = 11.84SASMSDD153 pKa = 3.47LDD155 pKa = 4.96EE156 pKa = 4.26PTQRR160 pKa = 11.84EE161 pKa = 4.21NSWRR165 pKa = 11.84RR166 pKa = 11.84QRR168 pKa = 11.84HH169 pKa = 4.34GPITAAMRR177 pKa = 11.84GSGRR181 pKa = 11.84GRR183 pKa = 11.84GRR185 pKa = 11.84GQGPRR190 pKa = 11.84MGYY193 pKa = 9.83RR194 pKa = 11.84PGGVGGNEE202 pKa = 3.72NGGGVFYY209 pKa = 11.17SLGHH213 pKa = 4.82VV214 pKa = 3.47
MM1 pKa = 7.4VYY3 pKa = 10.67YY4 pKa = 10.08ILLLGLVGLGLGEE17 pKa = 4.3STNTSSSNSSSIASSLPPPKK37 pKa = 8.85ITHH40 pKa = 6.47IPTTTSPTVTVTVTDD55 pKa = 3.98LMLGLVVLLLVVILSLLVVILLTLKK80 pKa = 10.64RR81 pKa = 11.84MRR83 pKa = 11.84KK84 pKa = 8.85DD85 pKa = 3.09AKK87 pKa = 10.71AEE89 pKa = 3.82KK90 pKa = 10.08DD91 pKa = 3.52KK92 pKa = 11.15KK93 pKa = 10.55QKK95 pKa = 10.29PDD97 pKa = 3.69DD98 pKa = 3.86VEE100 pKa = 4.05MSLLRR105 pKa = 11.84RR106 pKa = 11.84MHH108 pKa = 6.77SDD110 pKa = 2.72HH111 pKa = 5.86MHH113 pKa = 5.21MHH115 pKa = 6.32SEE117 pKa = 3.82AAGPFGNMYY126 pKa = 8.94TRR128 pKa = 11.84PLPSPPQNQWWQNGTLGGRR147 pKa = 11.84SASMSDD153 pKa = 3.47LDD155 pKa = 4.96EE156 pKa = 4.26PTQRR160 pKa = 11.84EE161 pKa = 4.21NSWRR165 pKa = 11.84RR166 pKa = 11.84QRR168 pKa = 11.84HH169 pKa = 4.34GPITAAMRR177 pKa = 11.84GSGRR181 pKa = 11.84GRR183 pKa = 11.84GRR185 pKa = 11.84GQGPRR190 pKa = 11.84MGYY193 pKa = 9.83RR194 pKa = 11.84PGGVGGNEE202 pKa = 3.72NGGGVFYY209 pKa = 11.17SLGHH213 pKa = 4.82VV214 pKa = 3.47
Molecular weight: 23.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
78638 |
62 |
4113 |
524.3 |
58.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.735 ± 0.161 | 2.208 ± 0.102 |
6.062 ± 0.165 | 6.023 ± 0.21 |
3.559 ± 0.096 | 5.13 ± 0.168 |
2.173 ± 0.109 | 4.144 ± 0.088 |
5.366 ± 0.204 | 8.636 ± 0.17 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.809 ± 0.092 | 4.106 ± 0.159 |
5.42 ± 0.174 | 3.942 ± 0.174 |
5.605 ± 0.143 | 8.113 ± 0.185 |
7.405 ± 0.37 | 7.035 ± 0.113 |
1.283 ± 0.074 | 3.245 ± 0.098 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
