
Paracoccus aestuariivivens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
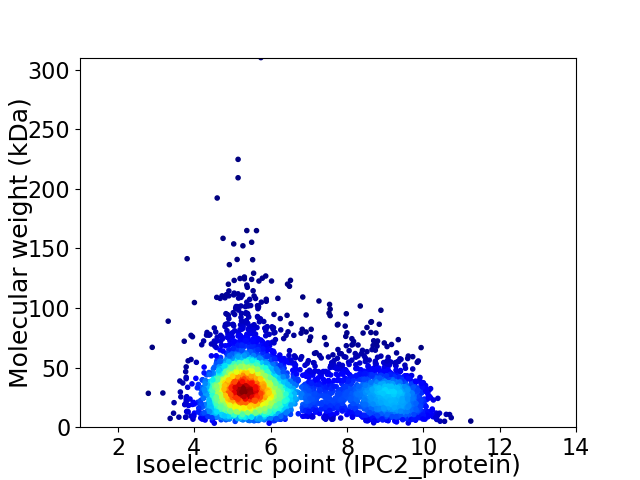
Virtual 2D-PAGE plot for 4256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L6J9Z4|A0A6L6J9Z4_9RHOB Glycosyltransferase OS=Paracoccus aestuariivivens OX=1820333 GN=GL286_13940 PE=4 SV=1
MM1 pKa = 8.23RR2 pKa = 11.84DD3 pKa = 3.23TSVPSSDD10 pKa = 4.96FMNDD14 pKa = 2.98PQTDD18 pKa = 3.68QSVSTSRR25 pKa = 11.84AIRR28 pKa = 11.84TEE30 pKa = 3.73TSDD33 pKa = 3.46ADD35 pKa = 3.7SGISTRR41 pKa = 11.84YY42 pKa = 7.35TMNVGDD48 pKa = 4.01TFVGNLATYY57 pKa = 10.75SDD59 pKa = 4.32NDD61 pKa = 3.62WVRR64 pKa = 11.84VTLTAGTYY72 pKa = 9.95EE73 pKa = 4.01IDD75 pKa = 3.61LAGSGSSPVRR85 pKa = 11.84DD86 pKa = 3.59TYY88 pKa = 11.56LRR90 pKa = 11.84IYY92 pKa = 10.31DD93 pKa = 3.62ATGTLVTYY101 pKa = 10.86NDD103 pKa = 4.47DD104 pKa = 4.56GGDD107 pKa = 3.66GLNSHH112 pKa = 7.29LSLTVTQPGTYY123 pKa = 8.9YY124 pKa = 10.66VSAGAFSSSYY134 pKa = 9.18TGSYY138 pKa = 9.63SLNVSRR144 pKa = 11.84GAEE147 pKa = 3.73PTVFTIAQIAAQLTDD162 pKa = 4.78GYY164 pKa = 10.33WDD166 pKa = 3.3STDD169 pKa = 3.11RR170 pKa = 11.84GRR172 pKa = 11.84RR173 pKa = 11.84SFDD176 pKa = 3.16VSPGEE181 pKa = 4.26TINVDD186 pKa = 4.47LSGLDD191 pKa = 3.17ADD193 pKa = 4.29GRR195 pKa = 11.84RR196 pKa = 11.84LAVAALAAWTDD207 pKa = 3.33VSGIRR212 pKa = 11.84FNSVNPYY219 pKa = 10.08SGQTVHH225 pKa = 5.99ITFGDD230 pKa = 3.94DD231 pKa = 3.89EE232 pKa = 5.21SGAWSSSTTSGGEE245 pKa = 3.46IWSSDD250 pKa = 3.62VNVGRR255 pKa = 11.84DD256 pKa = 2.94WLDD259 pKa = 3.12TYY261 pKa = 11.0GTGFATYY268 pKa = 10.01SYY270 pKa = 8.93QTYY273 pKa = 9.13IHH275 pKa = 7.16EE276 pKa = 4.71IGHH279 pKa = 6.42ALGLGHH285 pKa = 7.58AGNYY289 pKa = 9.08NGSAVYY295 pKa = 8.45GTDD298 pKa = 2.7NTYY301 pKa = 10.85INDD304 pKa = 3.2SWQASVMSYY313 pKa = 10.02FSQDD317 pKa = 2.89DD318 pKa = 3.73NTFVNASYY326 pKa = 10.46AYY328 pKa = 10.68VMSAMAADD336 pKa = 3.75IAAIQEE342 pKa = 4.32LYY344 pKa = 11.18GRR346 pKa = 11.84TTLRR350 pKa = 11.84AGNTVYY356 pKa = 11.06GEE358 pKa = 3.96NSNAGGNYY366 pKa = 9.51GRR368 pKa = 11.84ISSILATGARR378 pKa = 11.84DD379 pKa = 4.28DD380 pKa = 3.58IAFTIVDD387 pKa = 3.88SQGVDD392 pKa = 3.32TLNLSGDD399 pKa = 3.77TANQRR404 pKa = 11.84IIMTPGGISNAYY416 pKa = 9.39GITGNIIITSGTVIEE431 pKa = 4.23NLRR434 pKa = 11.84AGSGNDD440 pKa = 3.45YY441 pKa = 11.04LSGNAAHH448 pKa = 5.85NTIWGNAGNDD458 pKa = 3.7MIIGGLGNDD467 pKa = 3.95TLLGGAGRR475 pKa = 11.84DD476 pKa = 3.65TLNGGNGNDD485 pKa = 3.8IYY487 pKa = 11.03HH488 pKa = 7.48ADD490 pKa = 3.54GFDD493 pKa = 3.99IISEE497 pKa = 4.18GSNGGVDD504 pKa = 3.56TVYY507 pKa = 11.17SSGNFALTPNLEE519 pKa = 3.99NLYY522 pKa = 9.16LTGSAQTGTGNVAANTIVGNAQGNTLRR549 pKa = 11.84GMGGNDD555 pKa = 3.42ILNGAAGSDD564 pKa = 3.76TMHH567 pKa = 6.91GGAGNDD573 pKa = 3.51TYY575 pKa = 11.08HH576 pKa = 7.32IDD578 pKa = 3.53SQDD581 pKa = 3.45RR582 pKa = 11.84LVEE585 pKa = 3.98LANEE589 pKa = 4.96GIDD592 pKa = 3.65TVVSAASFTLAANFEE607 pKa = 4.29NLRR610 pKa = 11.84LTGSAVFGTGNDD622 pKa = 3.49AANTIVGNSLGNTIRR637 pKa = 11.84GMAGNDD643 pKa = 3.07ILNGGGGNDD652 pKa = 3.1RR653 pKa = 11.84LFGGTHH659 pKa = 6.95ADD661 pKa = 3.34TFVFGSGQDD670 pKa = 3.26MVMDD674 pKa = 4.08FQNNIDD680 pKa = 3.84TLRR683 pKa = 11.84IDD685 pKa = 3.54DD686 pKa = 4.63ALWGGGARR694 pKa = 11.84TVGQVLSLATVINGDD709 pKa = 3.85TVFLFGNGNRR719 pKa = 11.84LTIDD723 pKa = 2.97NYY725 pKa = 10.85TNINSLSDD733 pKa = 3.65DD734 pKa = 3.84LVIII738 pKa = 4.8
MM1 pKa = 8.23RR2 pKa = 11.84DD3 pKa = 3.23TSVPSSDD10 pKa = 4.96FMNDD14 pKa = 2.98PQTDD18 pKa = 3.68QSVSTSRR25 pKa = 11.84AIRR28 pKa = 11.84TEE30 pKa = 3.73TSDD33 pKa = 3.46ADD35 pKa = 3.7SGISTRR41 pKa = 11.84YY42 pKa = 7.35TMNVGDD48 pKa = 4.01TFVGNLATYY57 pKa = 10.75SDD59 pKa = 4.32NDD61 pKa = 3.62WVRR64 pKa = 11.84VTLTAGTYY72 pKa = 9.95EE73 pKa = 4.01IDD75 pKa = 3.61LAGSGSSPVRR85 pKa = 11.84DD86 pKa = 3.59TYY88 pKa = 11.56LRR90 pKa = 11.84IYY92 pKa = 10.31DD93 pKa = 3.62ATGTLVTYY101 pKa = 10.86NDD103 pKa = 4.47DD104 pKa = 4.56GGDD107 pKa = 3.66GLNSHH112 pKa = 7.29LSLTVTQPGTYY123 pKa = 8.9YY124 pKa = 10.66VSAGAFSSSYY134 pKa = 9.18TGSYY138 pKa = 9.63SLNVSRR144 pKa = 11.84GAEE147 pKa = 3.73PTVFTIAQIAAQLTDD162 pKa = 4.78GYY164 pKa = 10.33WDD166 pKa = 3.3STDD169 pKa = 3.11RR170 pKa = 11.84GRR172 pKa = 11.84RR173 pKa = 11.84SFDD176 pKa = 3.16VSPGEE181 pKa = 4.26TINVDD186 pKa = 4.47LSGLDD191 pKa = 3.17ADD193 pKa = 4.29GRR195 pKa = 11.84RR196 pKa = 11.84LAVAALAAWTDD207 pKa = 3.33VSGIRR212 pKa = 11.84FNSVNPYY219 pKa = 10.08SGQTVHH225 pKa = 5.99ITFGDD230 pKa = 3.94DD231 pKa = 3.89EE232 pKa = 5.21SGAWSSSTTSGGEE245 pKa = 3.46IWSSDD250 pKa = 3.62VNVGRR255 pKa = 11.84DD256 pKa = 2.94WLDD259 pKa = 3.12TYY261 pKa = 11.0GTGFATYY268 pKa = 10.01SYY270 pKa = 8.93QTYY273 pKa = 9.13IHH275 pKa = 7.16EE276 pKa = 4.71IGHH279 pKa = 6.42ALGLGHH285 pKa = 7.58AGNYY289 pKa = 9.08NGSAVYY295 pKa = 8.45GTDD298 pKa = 2.7NTYY301 pKa = 10.85INDD304 pKa = 3.2SWQASVMSYY313 pKa = 10.02FSQDD317 pKa = 2.89DD318 pKa = 3.73NTFVNASYY326 pKa = 10.46AYY328 pKa = 10.68VMSAMAADD336 pKa = 3.75IAAIQEE342 pKa = 4.32LYY344 pKa = 11.18GRR346 pKa = 11.84TTLRR350 pKa = 11.84AGNTVYY356 pKa = 11.06GEE358 pKa = 3.96NSNAGGNYY366 pKa = 9.51GRR368 pKa = 11.84ISSILATGARR378 pKa = 11.84DD379 pKa = 4.28DD380 pKa = 3.58IAFTIVDD387 pKa = 3.88SQGVDD392 pKa = 3.32TLNLSGDD399 pKa = 3.77TANQRR404 pKa = 11.84IIMTPGGISNAYY416 pKa = 9.39GITGNIIITSGTVIEE431 pKa = 4.23NLRR434 pKa = 11.84AGSGNDD440 pKa = 3.45YY441 pKa = 11.04LSGNAAHH448 pKa = 5.85NTIWGNAGNDD458 pKa = 3.7MIIGGLGNDD467 pKa = 3.95TLLGGAGRR475 pKa = 11.84DD476 pKa = 3.65TLNGGNGNDD485 pKa = 3.8IYY487 pKa = 11.03HH488 pKa = 7.48ADD490 pKa = 3.54GFDD493 pKa = 3.99IISEE497 pKa = 4.18GSNGGVDD504 pKa = 3.56TVYY507 pKa = 11.17SSGNFALTPNLEE519 pKa = 3.99NLYY522 pKa = 9.16LTGSAQTGTGNVAANTIVGNAQGNTLRR549 pKa = 11.84GMGGNDD555 pKa = 3.42ILNGAAGSDD564 pKa = 3.76TMHH567 pKa = 6.91GGAGNDD573 pKa = 3.51TYY575 pKa = 11.08HH576 pKa = 7.32IDD578 pKa = 3.53SQDD581 pKa = 3.45RR582 pKa = 11.84LVEE585 pKa = 3.98LANEE589 pKa = 4.96GIDD592 pKa = 3.65TVVSAASFTLAANFEE607 pKa = 4.29NLRR610 pKa = 11.84LTGSAVFGTGNDD622 pKa = 3.49AANTIVGNSLGNTIRR637 pKa = 11.84GMAGNDD643 pKa = 3.07ILNGGGGNDD652 pKa = 3.1RR653 pKa = 11.84LFGGTHH659 pKa = 6.95ADD661 pKa = 3.34TFVFGSGQDD670 pKa = 3.26MVMDD674 pKa = 4.08FQNNIDD680 pKa = 3.84TLRR683 pKa = 11.84IDD685 pKa = 3.54DD686 pKa = 4.63ALWGGGARR694 pKa = 11.84TVGQVLSLATVINGDD709 pKa = 3.85TVFLFGNGNRR719 pKa = 11.84LTIDD723 pKa = 2.97NYY725 pKa = 10.85TNINSLSDD733 pKa = 3.65DD734 pKa = 3.84LVIII738 pKa = 4.8
Molecular weight: 77.05 kDa
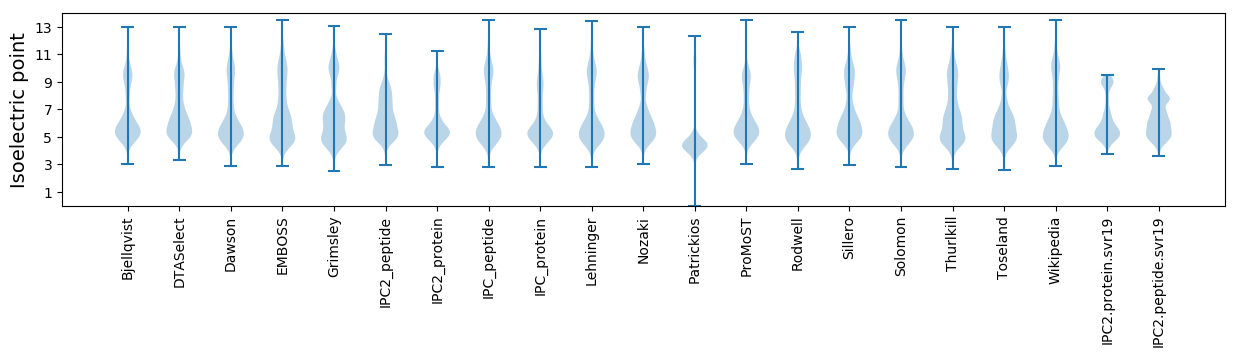
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L6J370|A0A6L6J370_9RHOB Uncharacterized protein OS=Paracoccus aestuariivivens OX=1820333 GN=GL286_02455 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 8.91SLSAA45 pKa = 3.86
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 8.91SLSAA45 pKa = 3.86
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1347648 |
29 |
2777 |
316.6 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.279 ± 0.04 | 0.815 ± 0.012 |
5.81 ± 0.032 | 5.605 ± 0.036 |
3.63 ± 0.025 | 8.667 ± 0.038 |
2.067 ± 0.019 | 5.494 ± 0.028 |
2.991 ± 0.03 | 10.331 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.786 ± 0.017 | 2.624 ± 0.02 |
5.229 ± 0.027 | 3.304 ± 0.021 |
6.992 ± 0.043 | 5.428 ± 0.027 |
5.282 ± 0.024 | 7.097 ± 0.025 |
1.431 ± 0.015 | 2.138 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
