
Beihai narna-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
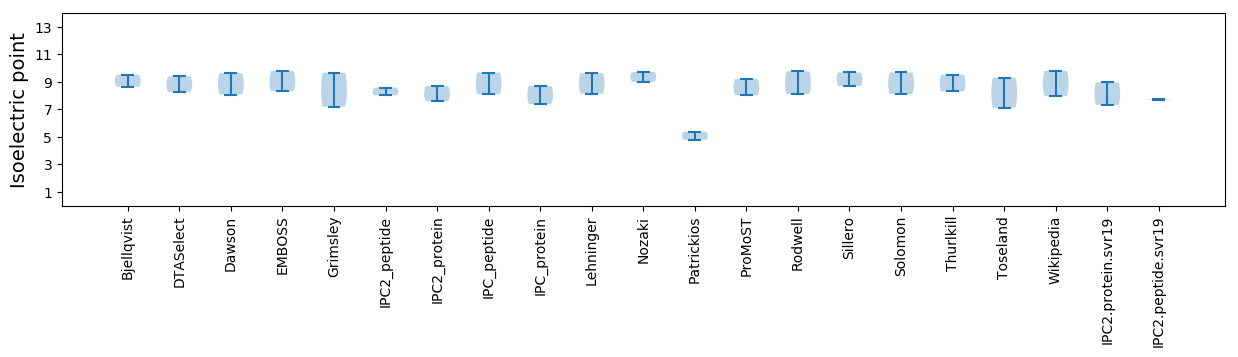
Average proteome isoelectric point is 8.15
Get precalculated fractions of proteins
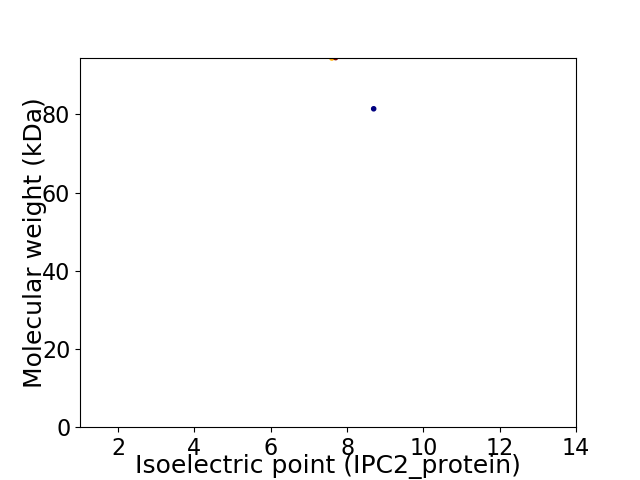
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI99|A0A1L3KI99_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 24 OX=1922452 PE=4 SV=1
MM1 pKa = 7.59ALVTDD6 pKa = 4.04WTTAVGRR13 pKa = 11.84LHH15 pKa = 7.59RR16 pKa = 11.84SPPKK20 pKa = 10.29CAGDD24 pKa = 3.64SALGRR29 pKa = 11.84TGDD32 pKa = 4.25PKK34 pKa = 11.12DD35 pKa = 3.76EE36 pKa = 4.59PLGGTTGGGPPGPCGPDD53 pKa = 3.52LLPTPPEE60 pKa = 3.97GRR62 pKa = 11.84HH63 pKa = 4.29WGLRR67 pKa = 11.84GDD69 pKa = 3.95HH70 pKa = 6.21GSLRR74 pKa = 11.84NRR76 pKa = 11.84EE77 pKa = 3.91ASQVLLEE84 pKa = 4.33HH85 pKa = 6.5LTYY88 pKa = 10.15STGDD92 pKa = 3.36LPGTLEE98 pKa = 4.2KK99 pKa = 10.6EE100 pKa = 4.31AGGKK104 pKa = 8.9RR105 pKa = 11.84VPVQDD110 pKa = 3.35EE111 pKa = 4.26QEE113 pKa = 4.34LVRR116 pKa = 11.84EE117 pKa = 4.32AHH119 pKa = 6.5CEE121 pKa = 3.67AVLGIVTEE129 pKa = 4.26VRR131 pKa = 11.84PAPHH135 pKa = 6.58EE136 pKa = 4.38RR137 pKa = 11.84DD138 pKa = 3.24PALPGGDD145 pKa = 3.75GPHH148 pKa = 6.74LKK150 pKa = 10.53LLARR154 pKa = 11.84VVGPHH159 pKa = 5.17VPVRR163 pKa = 11.84DD164 pKa = 4.22VPHH167 pKa = 6.51HH168 pKa = 6.01RR169 pKa = 11.84CPRR172 pKa = 11.84PGWIDD177 pKa = 3.33QILPGSGPQEE187 pKa = 4.58GGDD190 pKa = 3.96PPPEE194 pKa = 4.12PLRR197 pKa = 11.84EE198 pKa = 4.11SSTLWVPPHH207 pKa = 6.13GRR209 pKa = 11.84QPVPAQRR216 pKa = 11.84GRR218 pKa = 11.84RR219 pKa = 11.84VDD221 pKa = 3.44RR222 pKa = 11.84KK223 pKa = 7.77TAAPGPTMDD232 pKa = 5.4ARR234 pKa = 11.84MEE236 pKa = 4.5DD237 pKa = 4.0SQDD240 pKa = 3.3PPHH243 pKa = 6.91RR244 pKa = 11.84PGVARR249 pKa = 11.84LSTGCLHH256 pKa = 7.44AVRR259 pKa = 11.84HH260 pKa = 5.68SDD262 pKa = 3.43PYY264 pKa = 11.39GKK266 pKa = 10.3GLPVLRR272 pKa = 11.84EE273 pKa = 3.87VEE275 pKa = 4.27RR276 pKa = 11.84PNQATEE282 pKa = 3.8GHH284 pKa = 6.36KK285 pKa = 10.94GKK287 pKa = 9.04MVNPCAPPGVPRR299 pKa = 11.84GLLHH303 pKa = 6.65HH304 pKa = 7.3PGRR307 pKa = 11.84QEE309 pKa = 3.61VRR311 pKa = 11.84EE312 pKa = 4.28GSLPWPATIYY322 pKa = 9.8SSSVIVPHH330 pKa = 6.05LHH332 pKa = 7.35AEE334 pKa = 4.29AEE336 pKa = 4.34CLLRR340 pKa = 11.84EE341 pKa = 4.29HH342 pKa = 6.4EE343 pKa = 4.27VRR345 pKa = 11.84PRR347 pKa = 11.84LVVLASGEE355 pKa = 4.05LPPTGLHH362 pKa = 6.76DD363 pKa = 4.38ALVPLDD369 pKa = 4.32HH370 pKa = 7.56RR371 pKa = 11.84SGPNGRR377 pKa = 11.84EE378 pKa = 3.37IVPTYY383 pKa = 10.39RR384 pKa = 11.84RR385 pKa = 11.84AGGCGGKK392 pKa = 8.0TGQGDD397 pKa = 3.45VTRR400 pKa = 11.84NSRR403 pKa = 11.84RR404 pKa = 11.84PGPLGPVQVYY414 pKa = 7.71QAEE417 pKa = 4.33DD418 pKa = 3.52SPGRR422 pKa = 11.84RR423 pKa = 11.84QPHH426 pKa = 6.25HH427 pKa = 6.7NPPLVHH433 pKa = 6.47EE434 pKa = 5.15LGPVGHH440 pKa = 7.78LDD442 pKa = 4.15RR443 pKa = 11.84EE444 pKa = 4.76GADD447 pKa = 3.14QVGPNLVQMRR457 pKa = 11.84RR458 pKa = 11.84GASSGTAKK466 pKa = 10.56DD467 pKa = 3.86APDD470 pKa = 4.68DD471 pKa = 4.02GLAEE475 pKa = 4.24VVRR478 pKa = 11.84EE479 pKa = 4.04EE480 pKa = 3.82VGGRR484 pKa = 11.84RR485 pKa = 11.84EE486 pKa = 4.02VRR488 pKa = 11.84GHH490 pKa = 6.02RR491 pKa = 11.84KK492 pKa = 9.16VLEE495 pKa = 4.31EE496 pKa = 3.93PHH498 pKa = 6.92AGPTDD503 pKa = 3.7QTTHH507 pKa = 6.73RR508 pKa = 11.84PCMVTVQDD516 pKa = 4.42GLCPLVPPQSLEE528 pKa = 3.89EE529 pKa = 4.38PNAEE533 pKa = 4.03EE534 pKa = 4.1MSKK537 pKa = 9.45IHH539 pKa = 5.91QGPRR543 pKa = 11.84ALGDD547 pKa = 3.8YY548 pKa = 9.4PDD550 pKa = 4.53LSAAFRR556 pKa = 11.84QRR558 pKa = 11.84HH559 pKa = 5.84RR560 pKa = 11.84IDD562 pKa = 3.54CYY564 pKa = 11.29GPGEE568 pKa = 4.27LRR570 pKa = 11.84KK571 pKa = 9.57LHH573 pKa = 5.86QSLVTHH579 pKa = 6.18QALTADD585 pKa = 4.08DD586 pKa = 4.07VDD588 pKa = 3.72IAVCKK593 pKa = 10.23ALHH596 pKa = 6.13CVEE599 pKa = 5.35PLPQGLPFWLRR610 pKa = 11.84RR611 pKa = 11.84FADD614 pKa = 4.18RR615 pKa = 11.84VCGPSRR621 pKa = 11.84DD622 pKa = 3.74VYY624 pKa = 11.18GAGSKK629 pKa = 10.32PSLGPGHH636 pKa = 7.89LFTGATRR643 pKa = 11.84QPSGRR648 pKa = 11.84NNRR651 pKa = 11.84PRR653 pKa = 11.84EE654 pKa = 3.97EE655 pKa = 4.13AGRR658 pKa = 11.84PPARR662 pKa = 11.84PLFRR666 pKa = 11.84GVQNGVRR673 pKa = 11.84CGGWLGQLFTVLRR686 pKa = 11.84EE687 pKa = 4.26CCVTLAGTCLGKK699 pKa = 10.47GASNIRR705 pKa = 11.84EE706 pKa = 4.49LEE708 pKa = 4.17RR709 pKa = 11.84PAVPVDD715 pKa = 4.21DD716 pKa = 4.58RR717 pKa = 11.84PQRR720 pKa = 11.84PSEE723 pKa = 4.14VLGEE727 pKa = 4.21ATTPPARR734 pKa = 11.84NGDD737 pKa = 4.09RR738 pKa = 11.84LHH740 pKa = 6.97HH741 pKa = 6.29PQAQDD746 pKa = 2.92SRR748 pKa = 11.84PALEE752 pKa = 4.85GGEE755 pKa = 4.22CVIRR759 pKa = 11.84SRR761 pKa = 11.84EE762 pKa = 4.23GGCPNQPPRR771 pKa = 11.84QPGDD775 pKa = 3.25PPRR778 pKa = 11.84RR779 pKa = 11.84LSSGMFRR786 pKa = 11.84EE787 pKa = 4.34VKK789 pKa = 9.66PAEE792 pKa = 4.01LVGRR796 pKa = 11.84EE797 pKa = 3.93ASEE800 pKa = 4.29HH801 pKa = 5.6CTEE804 pKa = 5.81CPLQPDD810 pKa = 4.69AIGRR814 pKa = 11.84AGQLQARR821 pKa = 11.84EE822 pKa = 4.25EE823 pKa = 4.05VRR825 pKa = 11.84EE826 pKa = 4.01AFPLGIRR833 pKa = 11.84GRR835 pKa = 11.84LPGTRR840 pKa = 11.84AGTRR844 pKa = 11.84PLRR847 pKa = 11.84SVRR850 pKa = 11.84HH851 pKa = 5.78RR852 pKa = 11.84GFGEE856 pKa = 3.94KK857 pKa = 9.38TPEE860 pKa = 4.73AIGPARR866 pKa = 11.84PNRR869 pKa = 11.84SCVLQGG875 pKa = 3.32
MM1 pKa = 7.59ALVTDD6 pKa = 4.04WTTAVGRR13 pKa = 11.84LHH15 pKa = 7.59RR16 pKa = 11.84SPPKK20 pKa = 10.29CAGDD24 pKa = 3.64SALGRR29 pKa = 11.84TGDD32 pKa = 4.25PKK34 pKa = 11.12DD35 pKa = 3.76EE36 pKa = 4.59PLGGTTGGGPPGPCGPDD53 pKa = 3.52LLPTPPEE60 pKa = 3.97GRR62 pKa = 11.84HH63 pKa = 4.29WGLRR67 pKa = 11.84GDD69 pKa = 3.95HH70 pKa = 6.21GSLRR74 pKa = 11.84NRR76 pKa = 11.84EE77 pKa = 3.91ASQVLLEE84 pKa = 4.33HH85 pKa = 6.5LTYY88 pKa = 10.15STGDD92 pKa = 3.36LPGTLEE98 pKa = 4.2KK99 pKa = 10.6EE100 pKa = 4.31AGGKK104 pKa = 8.9RR105 pKa = 11.84VPVQDD110 pKa = 3.35EE111 pKa = 4.26QEE113 pKa = 4.34LVRR116 pKa = 11.84EE117 pKa = 4.32AHH119 pKa = 6.5CEE121 pKa = 3.67AVLGIVTEE129 pKa = 4.26VRR131 pKa = 11.84PAPHH135 pKa = 6.58EE136 pKa = 4.38RR137 pKa = 11.84DD138 pKa = 3.24PALPGGDD145 pKa = 3.75GPHH148 pKa = 6.74LKK150 pKa = 10.53LLARR154 pKa = 11.84VVGPHH159 pKa = 5.17VPVRR163 pKa = 11.84DD164 pKa = 4.22VPHH167 pKa = 6.51HH168 pKa = 6.01RR169 pKa = 11.84CPRR172 pKa = 11.84PGWIDD177 pKa = 3.33QILPGSGPQEE187 pKa = 4.58GGDD190 pKa = 3.96PPPEE194 pKa = 4.12PLRR197 pKa = 11.84EE198 pKa = 4.11SSTLWVPPHH207 pKa = 6.13GRR209 pKa = 11.84QPVPAQRR216 pKa = 11.84GRR218 pKa = 11.84RR219 pKa = 11.84VDD221 pKa = 3.44RR222 pKa = 11.84KK223 pKa = 7.77TAAPGPTMDD232 pKa = 5.4ARR234 pKa = 11.84MEE236 pKa = 4.5DD237 pKa = 4.0SQDD240 pKa = 3.3PPHH243 pKa = 6.91RR244 pKa = 11.84PGVARR249 pKa = 11.84LSTGCLHH256 pKa = 7.44AVRR259 pKa = 11.84HH260 pKa = 5.68SDD262 pKa = 3.43PYY264 pKa = 11.39GKK266 pKa = 10.3GLPVLRR272 pKa = 11.84EE273 pKa = 3.87VEE275 pKa = 4.27RR276 pKa = 11.84PNQATEE282 pKa = 3.8GHH284 pKa = 6.36KK285 pKa = 10.94GKK287 pKa = 9.04MVNPCAPPGVPRR299 pKa = 11.84GLLHH303 pKa = 6.65HH304 pKa = 7.3PGRR307 pKa = 11.84QEE309 pKa = 3.61VRR311 pKa = 11.84EE312 pKa = 4.28GSLPWPATIYY322 pKa = 9.8SSSVIVPHH330 pKa = 6.05LHH332 pKa = 7.35AEE334 pKa = 4.29AEE336 pKa = 4.34CLLRR340 pKa = 11.84EE341 pKa = 4.29HH342 pKa = 6.4EE343 pKa = 4.27VRR345 pKa = 11.84PRR347 pKa = 11.84LVVLASGEE355 pKa = 4.05LPPTGLHH362 pKa = 6.76DD363 pKa = 4.38ALVPLDD369 pKa = 4.32HH370 pKa = 7.56RR371 pKa = 11.84SGPNGRR377 pKa = 11.84EE378 pKa = 3.37IVPTYY383 pKa = 10.39RR384 pKa = 11.84RR385 pKa = 11.84AGGCGGKK392 pKa = 8.0TGQGDD397 pKa = 3.45VTRR400 pKa = 11.84NSRR403 pKa = 11.84RR404 pKa = 11.84PGPLGPVQVYY414 pKa = 7.71QAEE417 pKa = 4.33DD418 pKa = 3.52SPGRR422 pKa = 11.84RR423 pKa = 11.84QPHH426 pKa = 6.25HH427 pKa = 6.7NPPLVHH433 pKa = 6.47EE434 pKa = 5.15LGPVGHH440 pKa = 7.78LDD442 pKa = 4.15RR443 pKa = 11.84EE444 pKa = 4.76GADD447 pKa = 3.14QVGPNLVQMRR457 pKa = 11.84RR458 pKa = 11.84GASSGTAKK466 pKa = 10.56DD467 pKa = 3.86APDD470 pKa = 4.68DD471 pKa = 4.02GLAEE475 pKa = 4.24VVRR478 pKa = 11.84EE479 pKa = 4.04EE480 pKa = 3.82VGGRR484 pKa = 11.84RR485 pKa = 11.84EE486 pKa = 4.02VRR488 pKa = 11.84GHH490 pKa = 6.02RR491 pKa = 11.84KK492 pKa = 9.16VLEE495 pKa = 4.31EE496 pKa = 3.93PHH498 pKa = 6.92AGPTDD503 pKa = 3.7QTTHH507 pKa = 6.73RR508 pKa = 11.84PCMVTVQDD516 pKa = 4.42GLCPLVPPQSLEE528 pKa = 3.89EE529 pKa = 4.38PNAEE533 pKa = 4.03EE534 pKa = 4.1MSKK537 pKa = 9.45IHH539 pKa = 5.91QGPRR543 pKa = 11.84ALGDD547 pKa = 3.8YY548 pKa = 9.4PDD550 pKa = 4.53LSAAFRR556 pKa = 11.84QRR558 pKa = 11.84HH559 pKa = 5.84RR560 pKa = 11.84IDD562 pKa = 3.54CYY564 pKa = 11.29GPGEE568 pKa = 4.27LRR570 pKa = 11.84KK571 pKa = 9.57LHH573 pKa = 5.86QSLVTHH579 pKa = 6.18QALTADD585 pKa = 4.08DD586 pKa = 4.07VDD588 pKa = 3.72IAVCKK593 pKa = 10.23ALHH596 pKa = 6.13CVEE599 pKa = 5.35PLPQGLPFWLRR610 pKa = 11.84RR611 pKa = 11.84FADD614 pKa = 4.18RR615 pKa = 11.84VCGPSRR621 pKa = 11.84DD622 pKa = 3.74VYY624 pKa = 11.18GAGSKK629 pKa = 10.32PSLGPGHH636 pKa = 7.89LFTGATRR643 pKa = 11.84QPSGRR648 pKa = 11.84NNRR651 pKa = 11.84PRR653 pKa = 11.84EE654 pKa = 3.97EE655 pKa = 4.13AGRR658 pKa = 11.84PPARR662 pKa = 11.84PLFRR666 pKa = 11.84GVQNGVRR673 pKa = 11.84CGGWLGQLFTVLRR686 pKa = 11.84EE687 pKa = 4.26CCVTLAGTCLGKK699 pKa = 10.47GASNIRR705 pKa = 11.84EE706 pKa = 4.49LEE708 pKa = 4.17RR709 pKa = 11.84PAVPVDD715 pKa = 4.21DD716 pKa = 4.58RR717 pKa = 11.84PQRR720 pKa = 11.84PSEE723 pKa = 4.14VLGEE727 pKa = 4.21ATTPPARR734 pKa = 11.84NGDD737 pKa = 4.09RR738 pKa = 11.84LHH740 pKa = 6.97HH741 pKa = 6.29PQAQDD746 pKa = 2.92SRR748 pKa = 11.84PALEE752 pKa = 4.85GGEE755 pKa = 4.22CVIRR759 pKa = 11.84SRR761 pKa = 11.84EE762 pKa = 4.23GGCPNQPPRR771 pKa = 11.84QPGDD775 pKa = 3.25PPRR778 pKa = 11.84RR779 pKa = 11.84LSSGMFRR786 pKa = 11.84EE787 pKa = 4.34VKK789 pKa = 9.66PAEE792 pKa = 4.01LVGRR796 pKa = 11.84EE797 pKa = 3.93ASEE800 pKa = 4.29HH801 pKa = 5.6CTEE804 pKa = 5.81CPLQPDD810 pKa = 4.69AIGRR814 pKa = 11.84AGQLQARR821 pKa = 11.84EE822 pKa = 4.25EE823 pKa = 4.05VRR825 pKa = 11.84EE826 pKa = 4.01AFPLGIRR833 pKa = 11.84GRR835 pKa = 11.84LPGTRR840 pKa = 11.84AGTRR844 pKa = 11.84PLRR847 pKa = 11.84SVRR850 pKa = 11.84HH851 pKa = 5.78RR852 pKa = 11.84GFGEE856 pKa = 3.94KK857 pKa = 9.38TPEE860 pKa = 4.73AIGPARR866 pKa = 11.84PNRR869 pKa = 11.84SCVLQGG875 pKa = 3.32
Molecular weight: 94.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI99|A0A1L3KI99_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 24 OX=1922452 PE=4 SV=1
MM1 pKa = 7.25EE2 pKa = 5.46AVPVTSRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84CGFTKK16 pKa = 10.33YY17 pKa = 10.07FRR19 pKa = 11.84GSLRR23 pKa = 11.84SIIDD27 pKa = 3.23RR28 pKa = 11.84DD29 pKa = 3.66CRR31 pKa = 11.84ALQFSYY37 pKa = 10.69VGRR40 pKa = 11.84ALPEE44 pKa = 4.14ASPGQCDD51 pKa = 3.44AALTKK56 pKa = 10.51YY57 pKa = 10.2RR58 pKa = 11.84EE59 pKa = 4.31EE60 pKa = 4.22LSQPATTPDD69 pKa = 3.98SILDD73 pKa = 3.78SAEE76 pKa = 3.25EE77 pKa = 3.9WARR80 pKa = 11.84RR81 pKa = 11.84WASRR85 pKa = 11.84FLSGPVVPPTWLSGGACKK103 pKa = 9.68EE104 pKa = 3.93VPRR107 pKa = 11.84AKK109 pKa = 10.5GGLGTRR115 pKa = 11.84SVDD118 pKa = 3.26VARR121 pKa = 11.84RR122 pKa = 11.84ATDD125 pKa = 4.86AICEE129 pKa = 4.0PSEE132 pKa = 4.27PEE134 pKa = 3.74WEE136 pKa = 4.34SLRR139 pKa = 11.84EE140 pKa = 3.74RR141 pKa = 11.84LNAVEE146 pKa = 5.68GLTDD150 pKa = 3.89CDD152 pKa = 3.15VDD154 pKa = 3.78IVCRR158 pKa = 11.84EE159 pKa = 3.87RR160 pKa = 11.84LVSNQALVEE169 pKa = 4.01LSEE172 pKa = 4.41LPRR175 pKa = 11.84PIAVDD180 pKa = 3.83AMALPEE186 pKa = 4.28RR187 pKa = 11.84GGKK190 pKa = 8.85VRR192 pKa = 11.84IVTKK196 pKa = 10.47CPWALVYY203 pKa = 10.5LGHH206 pKa = 7.15FLRR209 pKa = 11.84VWLLEE214 pKa = 3.46GLRR217 pKa = 11.84RR218 pKa = 11.84DD219 pKa = 3.61EE220 pKa = 4.17RR221 pKa = 11.84TKK223 pKa = 10.94SVLDD227 pKa = 3.8GDD229 pKa = 4.34HH230 pKa = 6.71AGSVGGLVGGAGVRR244 pKa = 11.84LLEE247 pKa = 4.23HH248 pKa = 6.66LAVSADD254 pKa = 3.49LTAASDD260 pKa = 4.19LLPHH264 pKa = 6.94DD265 pKa = 4.75LCQAIVRR272 pKa = 11.84GVLRR276 pKa = 11.84GSRR279 pKa = 11.84TRR281 pKa = 11.84PAPHH285 pKa = 7.05LDD287 pKa = 3.88EE288 pKa = 4.58VWADD292 pKa = 4.12LIGPLTIQMPDD303 pKa = 2.57GSEE306 pKa = 4.0FVNQRR311 pKa = 11.84GIMMGLPTTWTILSLVHH328 pKa = 6.76LYY330 pKa = 8.44WAEE333 pKa = 3.93WAWASTVPRR342 pKa = 11.84HH343 pKa = 5.91ISLSGLPSAAPRR355 pKa = 11.84TAICGDD361 pKa = 3.81DD362 pKa = 4.58LAAVWPTSVVQRR374 pKa = 11.84YY375 pKa = 7.17EE376 pKa = 4.55SIVKK380 pKa = 10.05SCGGQFSAGKK390 pKa = 8.94HH391 pKa = 4.3YY392 pKa = 10.76KK393 pKa = 10.03SRR395 pKa = 11.84TYY397 pKa = 11.63LMFTEE402 pKa = 4.32EE403 pKa = 4.03AFRR406 pKa = 11.84LRR408 pKa = 11.84VEE410 pKa = 3.8MRR412 pKa = 11.84DD413 pKa = 3.17YY414 pKa = 10.36DD415 pKa = 3.82TRR417 pKa = 11.84RR418 pKa = 11.84VDD420 pKa = 3.12GRR422 pKa = 11.84RR423 pKa = 11.84PGEE426 pKa = 3.7ATLADD431 pKa = 5.02FLPSWMVEE439 pKa = 4.04EE440 pKa = 4.89PSRR443 pKa = 11.84DD444 pKa = 3.7SRR446 pKa = 11.84WCTGVDD452 pKa = 3.56HH453 pKa = 7.05LPLVPLRR460 pKa = 11.84GLVRR464 pKa = 11.84PLHH467 pKa = 5.93LPKK470 pKa = 10.42DD471 pKa = 4.11RR472 pKa = 11.84QPLPVWVAVPHH483 pKa = 5.9CVEE486 pKa = 4.35AACTQSGDD494 pKa = 3.12AGAVRR499 pKa = 11.84RR500 pKa = 11.84ILRR503 pKa = 11.84VLHH506 pKa = 6.07PGIHH510 pKa = 5.53RR511 pKa = 11.84WARR514 pKa = 11.84SRR516 pKa = 11.84GFAVNAPTSLGGYY529 pKa = 7.78GLPPVRR535 pKa = 11.84GDD537 pKa = 3.42PKK539 pKa = 10.32RR540 pKa = 11.84ARR542 pKa = 11.84LPKK545 pKa = 8.24WFRR548 pKa = 11.84WGISSLLRR556 pKa = 11.84ATPWKK561 pKa = 10.68DD562 pKa = 3.67LINPARR568 pKa = 11.84AWAPVMRR575 pKa = 11.84DD576 pKa = 2.81IPHH579 pKa = 7.05RR580 pKa = 11.84DD581 pKa = 3.03MGADD585 pKa = 3.28HH586 pKa = 7.26ASEE589 pKa = 4.06KK590 pKa = 10.81LKK592 pKa = 11.07VGAVAARR599 pKa = 11.84KK600 pKa = 9.83GRR602 pKa = 11.84VPFMGRR608 pKa = 11.84GADD611 pKa = 3.42LGYY614 pKa = 10.63DD615 pKa = 3.32AEE617 pKa = 4.65DD618 pKa = 3.71RR619 pKa = 11.84LTVRR623 pKa = 11.84LSNEE627 pKa = 3.6LLFILDD633 pKa = 4.01RR634 pKa = 11.84DD635 pKa = 4.17TLPSSFFLQGPRR647 pKa = 11.84KK648 pKa = 9.91VSGAVRR654 pKa = 11.84KK655 pKa = 8.26MFKK658 pKa = 9.16QHH660 pKa = 5.94LRR662 pKa = 11.84RR663 pKa = 11.84FPIPEE668 pKa = 4.13GSVVASEE675 pKa = 4.53TPMASLRR682 pKa = 11.84RR683 pKa = 11.84RR684 pKa = 11.84WEE686 pKa = 3.89EE687 pKa = 3.49VRR689 pKa = 11.84TARR692 pKa = 11.84SWWSSPRR699 pKa = 11.84GAAEE703 pKa = 4.02GFVLGISSSAKK714 pKa = 9.42RR715 pKa = 11.84AVASALGWASMEE727 pKa = 4.28PAHH730 pKa = 6.87SGGPVSDD737 pKa = 3.85
MM1 pKa = 7.25EE2 pKa = 5.46AVPVTSRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84CGFTKK16 pKa = 10.33YY17 pKa = 10.07FRR19 pKa = 11.84GSLRR23 pKa = 11.84SIIDD27 pKa = 3.23RR28 pKa = 11.84DD29 pKa = 3.66CRR31 pKa = 11.84ALQFSYY37 pKa = 10.69VGRR40 pKa = 11.84ALPEE44 pKa = 4.14ASPGQCDD51 pKa = 3.44AALTKK56 pKa = 10.51YY57 pKa = 10.2RR58 pKa = 11.84EE59 pKa = 4.31EE60 pKa = 4.22LSQPATTPDD69 pKa = 3.98SILDD73 pKa = 3.78SAEE76 pKa = 3.25EE77 pKa = 3.9WARR80 pKa = 11.84RR81 pKa = 11.84WASRR85 pKa = 11.84FLSGPVVPPTWLSGGACKK103 pKa = 9.68EE104 pKa = 3.93VPRR107 pKa = 11.84AKK109 pKa = 10.5GGLGTRR115 pKa = 11.84SVDD118 pKa = 3.26VARR121 pKa = 11.84RR122 pKa = 11.84ATDD125 pKa = 4.86AICEE129 pKa = 4.0PSEE132 pKa = 4.27PEE134 pKa = 3.74WEE136 pKa = 4.34SLRR139 pKa = 11.84EE140 pKa = 3.74RR141 pKa = 11.84LNAVEE146 pKa = 5.68GLTDD150 pKa = 3.89CDD152 pKa = 3.15VDD154 pKa = 3.78IVCRR158 pKa = 11.84EE159 pKa = 3.87RR160 pKa = 11.84LVSNQALVEE169 pKa = 4.01LSEE172 pKa = 4.41LPRR175 pKa = 11.84PIAVDD180 pKa = 3.83AMALPEE186 pKa = 4.28RR187 pKa = 11.84GGKK190 pKa = 8.85VRR192 pKa = 11.84IVTKK196 pKa = 10.47CPWALVYY203 pKa = 10.5LGHH206 pKa = 7.15FLRR209 pKa = 11.84VWLLEE214 pKa = 3.46GLRR217 pKa = 11.84RR218 pKa = 11.84DD219 pKa = 3.61EE220 pKa = 4.17RR221 pKa = 11.84TKK223 pKa = 10.94SVLDD227 pKa = 3.8GDD229 pKa = 4.34HH230 pKa = 6.71AGSVGGLVGGAGVRR244 pKa = 11.84LLEE247 pKa = 4.23HH248 pKa = 6.66LAVSADD254 pKa = 3.49LTAASDD260 pKa = 4.19LLPHH264 pKa = 6.94DD265 pKa = 4.75LCQAIVRR272 pKa = 11.84GVLRR276 pKa = 11.84GSRR279 pKa = 11.84TRR281 pKa = 11.84PAPHH285 pKa = 7.05LDD287 pKa = 3.88EE288 pKa = 4.58VWADD292 pKa = 4.12LIGPLTIQMPDD303 pKa = 2.57GSEE306 pKa = 4.0FVNQRR311 pKa = 11.84GIMMGLPTTWTILSLVHH328 pKa = 6.76LYY330 pKa = 8.44WAEE333 pKa = 3.93WAWASTVPRR342 pKa = 11.84HH343 pKa = 5.91ISLSGLPSAAPRR355 pKa = 11.84TAICGDD361 pKa = 3.81DD362 pKa = 4.58LAAVWPTSVVQRR374 pKa = 11.84YY375 pKa = 7.17EE376 pKa = 4.55SIVKK380 pKa = 10.05SCGGQFSAGKK390 pKa = 8.94HH391 pKa = 4.3YY392 pKa = 10.76KK393 pKa = 10.03SRR395 pKa = 11.84TYY397 pKa = 11.63LMFTEE402 pKa = 4.32EE403 pKa = 4.03AFRR406 pKa = 11.84LRR408 pKa = 11.84VEE410 pKa = 3.8MRR412 pKa = 11.84DD413 pKa = 3.17YY414 pKa = 10.36DD415 pKa = 3.82TRR417 pKa = 11.84RR418 pKa = 11.84VDD420 pKa = 3.12GRR422 pKa = 11.84RR423 pKa = 11.84PGEE426 pKa = 3.7ATLADD431 pKa = 5.02FLPSWMVEE439 pKa = 4.04EE440 pKa = 4.89PSRR443 pKa = 11.84DD444 pKa = 3.7SRR446 pKa = 11.84WCTGVDD452 pKa = 3.56HH453 pKa = 7.05LPLVPLRR460 pKa = 11.84GLVRR464 pKa = 11.84PLHH467 pKa = 5.93LPKK470 pKa = 10.42DD471 pKa = 4.11RR472 pKa = 11.84QPLPVWVAVPHH483 pKa = 5.9CVEE486 pKa = 4.35AACTQSGDD494 pKa = 3.12AGAVRR499 pKa = 11.84RR500 pKa = 11.84ILRR503 pKa = 11.84VLHH506 pKa = 6.07PGIHH510 pKa = 5.53RR511 pKa = 11.84WARR514 pKa = 11.84SRR516 pKa = 11.84GFAVNAPTSLGGYY529 pKa = 7.78GLPPVRR535 pKa = 11.84GDD537 pKa = 3.42PKK539 pKa = 10.32RR540 pKa = 11.84ARR542 pKa = 11.84LPKK545 pKa = 8.24WFRR548 pKa = 11.84WGISSLLRR556 pKa = 11.84ATPWKK561 pKa = 10.68DD562 pKa = 3.67LINPARR568 pKa = 11.84AWAPVMRR575 pKa = 11.84DD576 pKa = 2.81IPHH579 pKa = 7.05RR580 pKa = 11.84DD581 pKa = 3.03MGADD585 pKa = 3.28HH586 pKa = 7.26ASEE589 pKa = 4.06KK590 pKa = 10.81LKK592 pKa = 11.07VGAVAARR599 pKa = 11.84KK600 pKa = 9.83GRR602 pKa = 11.84VPFMGRR608 pKa = 11.84GADD611 pKa = 3.42LGYY614 pKa = 10.63DD615 pKa = 3.32AEE617 pKa = 4.65DD618 pKa = 3.71RR619 pKa = 11.84LTVRR623 pKa = 11.84LSNEE627 pKa = 3.6LLFILDD633 pKa = 4.01RR634 pKa = 11.84DD635 pKa = 4.17TLPSSFFLQGPRR647 pKa = 11.84KK648 pKa = 9.91VSGAVRR654 pKa = 11.84KK655 pKa = 8.26MFKK658 pKa = 9.16QHH660 pKa = 5.94LRR662 pKa = 11.84RR663 pKa = 11.84FPIPEE668 pKa = 4.13GSVVASEE675 pKa = 4.53TPMASLRR682 pKa = 11.84RR683 pKa = 11.84RR684 pKa = 11.84WEE686 pKa = 3.89EE687 pKa = 3.49VRR689 pKa = 11.84TARR692 pKa = 11.84SWWSSPRR699 pKa = 11.84GAAEE703 pKa = 4.02GFVLGISSSAKK714 pKa = 9.42RR715 pKa = 11.84AVASALGWASMEE727 pKa = 4.28PAHH730 pKa = 6.87SGGPVSDD737 pKa = 3.85
Molecular weight: 81.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1612 |
737 |
875 |
806.0 |
87.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.375 ± 1.162 | 2.295 ± 0.276 |
5.211 ± 0.246 | 6.328 ± 0.628 |
1.737 ± 0.586 | 10.422 ± 1.496 |
3.474 ± 0.814 | 2.295 ± 0.576 |
2.481 ± 0.257 | 9.491 ± 0.478 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.365 ± 0.373 | 1.303 ± 0.341 |
10.112 ± 1.942 | 3.04 ± 0.89 |
10.856 ± 0.001 | 5.955 ± 1.335 |
4.404 ± 0.044 | 7.754 ± 0.27 |
1.923 ± 0.93 | 1.179 ± 0.219 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
