
Raccoon polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Procyon lotor polyomavirus 1
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
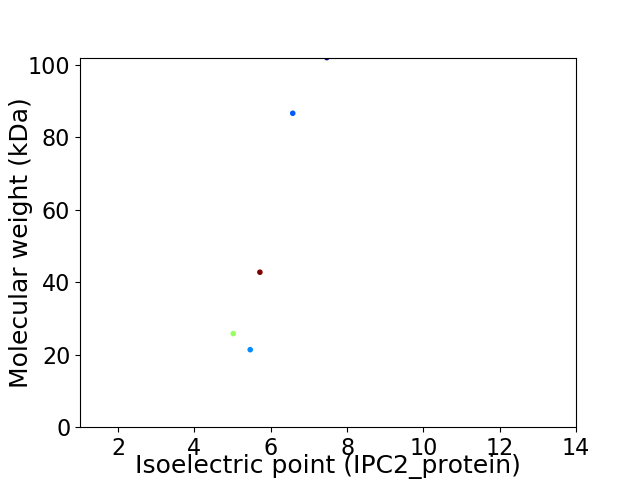
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7FZP3|J7FZP3_9POLY Small T antigen OS=Raccoon polyomavirus OX=1219896 PE=4 SV=1
MM1 pKa = 7.61GGFLSLFLSLGEE13 pKa = 4.12IVAEE17 pKa = 4.54LSATTGIAVEE27 pKa = 4.59SILTGEE33 pKa = 4.08ALAAIEE39 pKa = 4.54AEE41 pKa = 4.55VGSLMTVEE49 pKa = 4.24GLSGIEE55 pKa = 4.24ALSQLGFTAEE65 pKa = 3.99QFSNFSLVSSLVNQAAGYY83 pKa = 10.27GVFAQTLSGISALVGAGIKK102 pKa = 10.05LALDD106 pKa = 3.54EE107 pKa = 4.36QSTVNHH113 pKa = 6.32NLYY116 pKa = 10.61SLSAAKK122 pKa = 10.27RR123 pKa = 11.84EE124 pKa = 4.06EE125 pKa = 3.82EE126 pKa = 3.73RR127 pKa = 11.84LVNNGIQKK135 pKa = 10.3LSWSFGLDD143 pKa = 3.29PLNWDD148 pKa = 3.11QSLLHH153 pKa = 6.59AVGKK157 pKa = 8.74SVWEE161 pKa = 4.2GLQKK165 pKa = 10.8HH166 pKa = 6.3PGVNHH171 pKa = 6.27VYY173 pKa = 11.03SNLEE177 pKa = 3.55ALLRR181 pKa = 11.84QARR184 pKa = 11.84WVVKK188 pKa = 10.49RR189 pKa = 11.84EE190 pKa = 4.06STSALPHH197 pKa = 5.85QIGEE201 pKa = 4.29VIHH204 pKa = 6.24FFEE207 pKa = 4.89SHH209 pKa = 4.13VHH211 pKa = 5.79SNGVPDD217 pKa = 3.82WLLPLILGLKK227 pKa = 9.78GDD229 pKa = 4.26PTPAWHH235 pKa = 6.77HH236 pKa = 6.66LEE238 pKa = 4.89DD239 pKa = 4.73GPP241 pKa = 5.13
MM1 pKa = 7.61GGFLSLFLSLGEE13 pKa = 4.12IVAEE17 pKa = 4.54LSATTGIAVEE27 pKa = 4.59SILTGEE33 pKa = 4.08ALAAIEE39 pKa = 4.54AEE41 pKa = 4.55VGSLMTVEE49 pKa = 4.24GLSGIEE55 pKa = 4.24ALSQLGFTAEE65 pKa = 3.99QFSNFSLVSSLVNQAAGYY83 pKa = 10.27GVFAQTLSGISALVGAGIKK102 pKa = 10.05LALDD106 pKa = 3.54EE107 pKa = 4.36QSTVNHH113 pKa = 6.32NLYY116 pKa = 10.61SLSAAKK122 pKa = 10.27RR123 pKa = 11.84EE124 pKa = 4.06EE125 pKa = 3.82EE126 pKa = 3.73RR127 pKa = 11.84LVNNGIQKK135 pKa = 10.3LSWSFGLDD143 pKa = 3.29PLNWDD148 pKa = 3.11QSLLHH153 pKa = 6.59AVGKK157 pKa = 8.74SVWEE161 pKa = 4.2GLQKK165 pKa = 10.8HH166 pKa = 6.3PGVNHH171 pKa = 6.27VYY173 pKa = 11.03SNLEE177 pKa = 3.55ALLRR181 pKa = 11.84QARR184 pKa = 11.84WVVKK188 pKa = 10.49RR189 pKa = 11.84EE190 pKa = 4.06STSALPHH197 pKa = 5.85QIGEE201 pKa = 4.29VIHH204 pKa = 6.24FFEE207 pKa = 4.89SHH209 pKa = 4.13VHH211 pKa = 5.79SNGVPDD217 pKa = 3.82WLLPLILGLKK227 pKa = 9.78GDD229 pKa = 4.26PTPAWHH235 pKa = 6.77HH236 pKa = 6.66LEE238 pKa = 4.89DD239 pKa = 4.73GPP241 pKa = 5.13
Molecular weight: 25.83 kDa
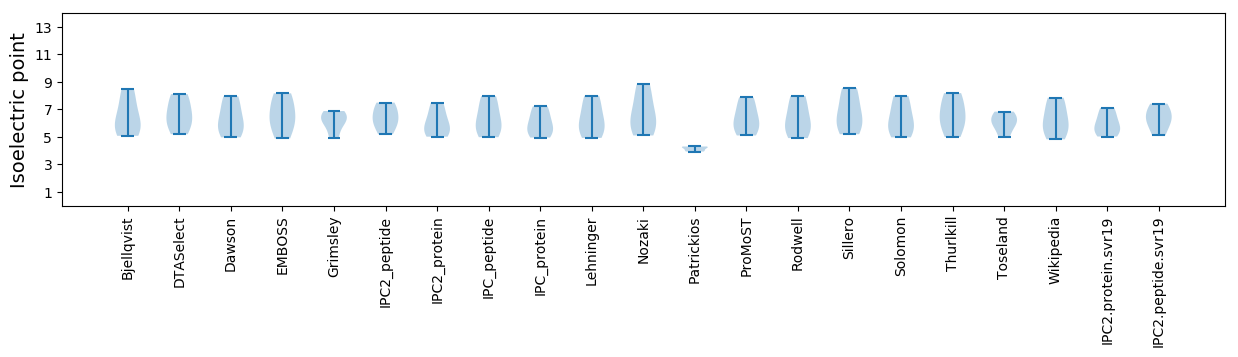
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7G293|J7G293_9POLY Capsid protein VP1 OS=Raccoon polyomavirus OX=1219896 PE=3 SV=1
MM1 pKa = 7.61DD2 pKa = 3.8TVLTSSEE9 pKa = 3.81RR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 4.07LCSLLEE18 pKa = 5.16LDD20 pKa = 3.79PQLWGNIPVMRR31 pKa = 11.84NCYY34 pKa = 9.85KK35 pKa = 10.56KK36 pKa = 10.66SCLKK40 pKa = 9.51YY41 pKa = 10.58HH42 pKa = 7.1PDD44 pKa = 4.22KK45 pKa = 11.65GGDD48 pKa = 3.52GQKK51 pKa = 9.18MVRR54 pKa = 11.84LNEE57 pKa = 3.53LWARR61 pKa = 11.84ATEE64 pKa = 3.93NLTRR68 pKa = 11.84LRR70 pKa = 11.84EE71 pKa = 4.15DD72 pKa = 3.64CGFQVSDD79 pKa = 4.57PLEE82 pKa = 4.52SSPKK86 pKa = 9.86TLMAHH91 pKa = 7.39LGSTWKK97 pKa = 10.76SKK99 pKa = 9.5FCKK102 pKa = 10.61GPTCRR107 pKa = 11.84INMKK111 pKa = 9.52PAYY114 pKa = 8.95CSCITCRR121 pKa = 11.84LYY123 pKa = 10.72KK124 pKa = 9.79QHH126 pKa = 6.58QAIKK130 pKa = 9.22KK131 pKa = 8.1KK132 pKa = 10.25KK133 pKa = 9.61KK134 pKa = 9.54RR135 pKa = 11.84RR136 pKa = 11.84CLVWGQCFCFSCYY149 pKa = 10.08LHH151 pKa = 6.89WFGFPNTWCFFDD163 pKa = 2.76WWMTLILEE171 pKa = 4.46TDD173 pKa = 3.75LNLLNLKK180 pKa = 8.94FCKK183 pKa = 10.57YY184 pKa = 10.2EE185 pKa = 3.71FLNIKK190 pKa = 9.42NFPYY194 pKa = 10.63QNASLMTFSLQDD206 pKa = 3.31EE207 pKa = 4.47PPIYY211 pKa = 8.89GTPAFRR217 pKa = 11.84EE218 pKa = 3.82WWFYY222 pKa = 8.63QQNLYY227 pKa = 8.28TWSHH231 pKa = 7.2DD232 pKa = 3.76GPSTSSSSGRR242 pKa = 11.84SRR244 pKa = 11.84RR245 pKa = 11.84WDD247 pKa = 3.09SSRR250 pKa = 11.84DD251 pKa = 3.33HH252 pKa = 7.56GEE254 pKa = 3.7FAGGEE259 pKa = 4.31GSSGRR264 pKa = 11.84DD265 pKa = 2.49RR266 pKa = 11.84VHH268 pKa = 6.27TSKK271 pKa = 10.91SSTSMPSHH279 pKa = 6.75RR280 pKa = 11.84PHH282 pKa = 7.61SSANLHH288 pKa = 6.43GDD290 pKa = 3.19NSEE293 pKa = 4.76DD294 pKa = 3.56PLFCEE299 pKa = 4.55EE300 pKa = 4.79TLDD303 pKa = 4.59SSPSPPPSPAADD315 pKa = 3.29PHH317 pKa = 6.39TFTSSQSSTPSSAFGFSSQTSTPSNASQGDD347 pKa = 3.73SRR349 pKa = 11.84TTGPHH354 pKa = 6.82PNTPSPRR361 pKa = 11.84RR362 pKa = 11.84RR363 pKa = 11.84KK364 pKa = 9.68HH365 pKa = 6.68RR366 pKa = 11.84EE367 pKa = 3.62DD368 pKa = 4.32HH369 pKa = 6.03PTSPYY374 pKa = 10.89ASGGSFSSTPPKK386 pKa = 10.22SKK388 pKa = 10.48KK389 pKa = 9.75PRR391 pKa = 11.84DD392 pKa = 3.68SFSPLDD398 pKa = 3.86FPSDD402 pKa = 3.92LLDD405 pKa = 3.97CLSRR409 pKa = 11.84ATLSNKK415 pKa = 6.74TQNAFLIFSTKK426 pKa = 10.25EE427 pKa = 4.09KK428 pKa = 11.02VCLLYY433 pKa = 10.96DD434 pKa = 3.69KK435 pKa = 10.93TDD437 pKa = 3.34DD438 pKa = 3.64KK439 pKa = 11.77YY440 pKa = 11.15KK441 pKa = 10.97VEE443 pKa = 4.75FKK445 pKa = 10.83SLHH448 pKa = 5.95KK449 pKa = 10.4YY450 pKa = 10.31KK451 pKa = 10.69NGSGFLFIILLTKK464 pKa = 10.51HH465 pKa = 5.81RR466 pKa = 11.84VTAIKK471 pKa = 10.31NFCAQFCTISFLLCKK486 pKa = 10.32AVIQPVQCYY495 pKa = 10.44DD496 pKa = 3.88RR497 pKa = 11.84LCKK500 pKa = 10.45DD501 pKa = 3.5PYY503 pKa = 11.04TFLKK507 pKa = 10.72ANKK510 pKa = 9.24CGIFEE515 pKa = 4.87GEE517 pKa = 4.05FAEE520 pKa = 5.58QKK522 pKa = 11.05DD523 pKa = 4.08PTCDD527 pKa = 2.81WNQIADD533 pKa = 3.69FALAHH538 pKa = 6.57LLDD541 pKa = 5.23DD542 pKa = 4.75PLLILAHH549 pKa = 6.32YY550 pKa = 10.43LDD552 pKa = 4.75FSKK555 pKa = 11.0PPGNCTKK562 pKa = 10.69CLTGKK567 pKa = 9.6GLKK570 pKa = 9.81AHH572 pKa = 7.12ADD574 pKa = 3.28HH575 pKa = 7.36AKK577 pKa = 10.06EE578 pKa = 3.97HH579 pKa = 7.27DD580 pKa = 3.61NAKK583 pKa = 10.59LFIKK587 pKa = 10.53SKK589 pKa = 7.7SQKK592 pKa = 10.37SICQQAADD600 pKa = 3.91VVCAKK605 pKa = 10.26KK606 pKa = 10.33RR607 pKa = 11.84LAMLEE612 pKa = 3.9QTRR615 pKa = 11.84TEE617 pKa = 3.83MLEE620 pKa = 3.91EE621 pKa = 4.21KK622 pKa = 10.06FRR624 pKa = 11.84KK625 pKa = 9.03QLAILKK631 pKa = 8.53EE632 pKa = 4.01LEE634 pKa = 4.42VIDD637 pKa = 4.12LHH639 pKa = 7.72LILGGVAWYY648 pKa = 10.89SVMFQNFADD657 pKa = 3.85MLKK660 pKa = 10.99GLLQLFTDD668 pKa = 5.11NIPKK672 pKa = 9.87KK673 pKa = 10.48RR674 pKa = 11.84NALFIGPINSGKK686 pKa = 9.01TSFAAALIDD695 pKa = 4.06LLEE698 pKa = 4.55GKK700 pKa = 9.48ALNINCPADD709 pKa = 3.44KK710 pKa = 11.2LNFEE714 pKa = 5.12LGCALDD720 pKa = 4.22LFMVCFEE727 pKa = 4.54DD728 pKa = 4.66VKK730 pKa = 11.12GSKK733 pKa = 10.18CSNKK737 pKa = 9.93DD738 pKa = 3.46LQSGQGMNNLDD749 pKa = 3.96NLRR752 pKa = 11.84DD753 pKa = 3.82HH754 pKa = 7.19MDD756 pKa = 3.48GAVPVQLEE764 pKa = 4.09RR765 pKa = 11.84KK766 pKa = 7.96HH767 pKa = 5.02QNKK770 pKa = 9.65RR771 pKa = 11.84SQIFPPCIVTCNEE784 pKa = 3.8YY785 pKa = 10.82YY786 pKa = 10.22IPGTVATRR794 pKa = 11.84FAQTLTFAIKK804 pKa = 10.53LCLKK808 pKa = 10.77NSIEE812 pKa = 4.62KK813 pKa = 10.31NILMQQKK820 pKa = 9.41RR821 pKa = 11.84VLQRR825 pKa = 11.84GCTLLMALIWLLPLDD840 pKa = 4.91DD841 pKa = 4.97FSDD844 pKa = 5.29GIRR847 pKa = 11.84SDD849 pKa = 3.65VKK851 pKa = 10.53DD852 pKa = 3.94FKK854 pKa = 11.38GAVEE858 pKa = 4.68GDD860 pKa = 4.03VGWATYY866 pKa = 10.76QKK868 pKa = 9.69MVEE871 pKa = 4.15NVEE874 pKa = 4.54NGDD877 pKa = 4.04DD878 pKa = 3.7PMKK881 pKa = 11.03GILEE885 pKa = 4.03QDD887 pKa = 3.35NEE889 pKa = 4.16QDD891 pKa = 4.13NEE893 pKa = 4.29DD894 pKa = 3.62VDD896 pKa = 4.31SGRR899 pKa = 11.84FTQQ902 pKa = 4.71
MM1 pKa = 7.61DD2 pKa = 3.8TVLTSSEE9 pKa = 3.81RR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 4.07LCSLLEE18 pKa = 5.16LDD20 pKa = 3.79PQLWGNIPVMRR31 pKa = 11.84NCYY34 pKa = 9.85KK35 pKa = 10.56KK36 pKa = 10.66SCLKK40 pKa = 9.51YY41 pKa = 10.58HH42 pKa = 7.1PDD44 pKa = 4.22KK45 pKa = 11.65GGDD48 pKa = 3.52GQKK51 pKa = 9.18MVRR54 pKa = 11.84LNEE57 pKa = 3.53LWARR61 pKa = 11.84ATEE64 pKa = 3.93NLTRR68 pKa = 11.84LRR70 pKa = 11.84EE71 pKa = 4.15DD72 pKa = 3.64CGFQVSDD79 pKa = 4.57PLEE82 pKa = 4.52SSPKK86 pKa = 9.86TLMAHH91 pKa = 7.39LGSTWKK97 pKa = 10.76SKK99 pKa = 9.5FCKK102 pKa = 10.61GPTCRR107 pKa = 11.84INMKK111 pKa = 9.52PAYY114 pKa = 8.95CSCITCRR121 pKa = 11.84LYY123 pKa = 10.72KK124 pKa = 9.79QHH126 pKa = 6.58QAIKK130 pKa = 9.22KK131 pKa = 8.1KK132 pKa = 10.25KK133 pKa = 9.61KK134 pKa = 9.54RR135 pKa = 11.84RR136 pKa = 11.84CLVWGQCFCFSCYY149 pKa = 10.08LHH151 pKa = 6.89WFGFPNTWCFFDD163 pKa = 2.76WWMTLILEE171 pKa = 4.46TDD173 pKa = 3.75LNLLNLKK180 pKa = 8.94FCKK183 pKa = 10.57YY184 pKa = 10.2EE185 pKa = 3.71FLNIKK190 pKa = 9.42NFPYY194 pKa = 10.63QNASLMTFSLQDD206 pKa = 3.31EE207 pKa = 4.47PPIYY211 pKa = 8.89GTPAFRR217 pKa = 11.84EE218 pKa = 3.82WWFYY222 pKa = 8.63QQNLYY227 pKa = 8.28TWSHH231 pKa = 7.2DD232 pKa = 3.76GPSTSSSSGRR242 pKa = 11.84SRR244 pKa = 11.84RR245 pKa = 11.84WDD247 pKa = 3.09SSRR250 pKa = 11.84DD251 pKa = 3.33HH252 pKa = 7.56GEE254 pKa = 3.7FAGGEE259 pKa = 4.31GSSGRR264 pKa = 11.84DD265 pKa = 2.49RR266 pKa = 11.84VHH268 pKa = 6.27TSKK271 pKa = 10.91SSTSMPSHH279 pKa = 6.75RR280 pKa = 11.84PHH282 pKa = 7.61SSANLHH288 pKa = 6.43GDD290 pKa = 3.19NSEE293 pKa = 4.76DD294 pKa = 3.56PLFCEE299 pKa = 4.55EE300 pKa = 4.79TLDD303 pKa = 4.59SSPSPPPSPAADD315 pKa = 3.29PHH317 pKa = 6.39TFTSSQSSTPSSAFGFSSQTSTPSNASQGDD347 pKa = 3.73SRR349 pKa = 11.84TTGPHH354 pKa = 6.82PNTPSPRR361 pKa = 11.84RR362 pKa = 11.84RR363 pKa = 11.84KK364 pKa = 9.68HH365 pKa = 6.68RR366 pKa = 11.84EE367 pKa = 3.62DD368 pKa = 4.32HH369 pKa = 6.03PTSPYY374 pKa = 10.89ASGGSFSSTPPKK386 pKa = 10.22SKK388 pKa = 10.48KK389 pKa = 9.75PRR391 pKa = 11.84DD392 pKa = 3.68SFSPLDD398 pKa = 3.86FPSDD402 pKa = 3.92LLDD405 pKa = 3.97CLSRR409 pKa = 11.84ATLSNKK415 pKa = 6.74TQNAFLIFSTKK426 pKa = 10.25EE427 pKa = 4.09KK428 pKa = 11.02VCLLYY433 pKa = 10.96DD434 pKa = 3.69KK435 pKa = 10.93TDD437 pKa = 3.34DD438 pKa = 3.64KK439 pKa = 11.77YY440 pKa = 11.15KK441 pKa = 10.97VEE443 pKa = 4.75FKK445 pKa = 10.83SLHH448 pKa = 5.95KK449 pKa = 10.4YY450 pKa = 10.31KK451 pKa = 10.69NGSGFLFIILLTKK464 pKa = 10.51HH465 pKa = 5.81RR466 pKa = 11.84VTAIKK471 pKa = 10.31NFCAQFCTISFLLCKK486 pKa = 10.32AVIQPVQCYY495 pKa = 10.44DD496 pKa = 3.88RR497 pKa = 11.84LCKK500 pKa = 10.45DD501 pKa = 3.5PYY503 pKa = 11.04TFLKK507 pKa = 10.72ANKK510 pKa = 9.24CGIFEE515 pKa = 4.87GEE517 pKa = 4.05FAEE520 pKa = 5.58QKK522 pKa = 11.05DD523 pKa = 4.08PTCDD527 pKa = 2.81WNQIADD533 pKa = 3.69FALAHH538 pKa = 6.57LLDD541 pKa = 5.23DD542 pKa = 4.75PLLILAHH549 pKa = 6.32YY550 pKa = 10.43LDD552 pKa = 4.75FSKK555 pKa = 11.0PPGNCTKK562 pKa = 10.69CLTGKK567 pKa = 9.6GLKK570 pKa = 9.81AHH572 pKa = 7.12ADD574 pKa = 3.28HH575 pKa = 7.36AKK577 pKa = 10.06EE578 pKa = 3.97HH579 pKa = 7.27DD580 pKa = 3.61NAKK583 pKa = 10.59LFIKK587 pKa = 10.53SKK589 pKa = 7.7SQKK592 pKa = 10.37SICQQAADD600 pKa = 3.91VVCAKK605 pKa = 10.26KK606 pKa = 10.33RR607 pKa = 11.84LAMLEE612 pKa = 3.9QTRR615 pKa = 11.84TEE617 pKa = 3.83MLEE620 pKa = 3.91EE621 pKa = 4.21KK622 pKa = 10.06FRR624 pKa = 11.84KK625 pKa = 9.03QLAILKK631 pKa = 8.53EE632 pKa = 4.01LEE634 pKa = 4.42VIDD637 pKa = 4.12LHH639 pKa = 7.72LILGGVAWYY648 pKa = 10.89SVMFQNFADD657 pKa = 3.85MLKK660 pKa = 10.99GLLQLFTDD668 pKa = 5.11NIPKK672 pKa = 9.87KK673 pKa = 10.48RR674 pKa = 11.84NALFIGPINSGKK686 pKa = 9.01TSFAAALIDD695 pKa = 4.06LLEE698 pKa = 4.55GKK700 pKa = 9.48ALNINCPADD709 pKa = 3.44KK710 pKa = 11.2LNFEE714 pKa = 5.12LGCALDD720 pKa = 4.22LFMVCFEE727 pKa = 4.54DD728 pKa = 4.66VKK730 pKa = 11.12GSKK733 pKa = 10.18CSNKK737 pKa = 9.93DD738 pKa = 3.46LQSGQGMNNLDD749 pKa = 3.96NLRR752 pKa = 11.84DD753 pKa = 3.82HH754 pKa = 7.19MDD756 pKa = 3.48GAVPVQLEE764 pKa = 4.09RR765 pKa = 11.84KK766 pKa = 7.96HH767 pKa = 5.02QNKK770 pKa = 9.65RR771 pKa = 11.84SQIFPPCIVTCNEE784 pKa = 3.8YY785 pKa = 10.82YY786 pKa = 10.22IPGTVATRR794 pKa = 11.84FAQTLTFAIKK804 pKa = 10.53LCLKK808 pKa = 10.77NSIEE812 pKa = 4.62KK813 pKa = 10.31NILMQQKK820 pKa = 9.41RR821 pKa = 11.84VLQRR825 pKa = 11.84GCTLLMALIWLLPLDD840 pKa = 4.91DD841 pKa = 4.97FSDD844 pKa = 5.29GIRR847 pKa = 11.84SDD849 pKa = 3.65VKK851 pKa = 10.53DD852 pKa = 3.94FKK854 pKa = 11.38GAVEE858 pKa = 4.68GDD860 pKa = 4.03VGWATYY866 pKa = 10.76QKK868 pKa = 9.69MVEE871 pKa = 4.15NVEE874 pKa = 4.54NGDD877 pKa = 4.04DD878 pKa = 3.7PMKK881 pKa = 11.03GILEE885 pKa = 4.03QDD887 pKa = 3.35NEE889 pKa = 4.16QDD891 pKa = 4.13NEE893 pKa = 4.29DD894 pKa = 3.62VDD896 pKa = 4.31SGRR899 pKa = 11.84FTQQ902 pKa = 4.71
Molecular weight: 102.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2496 |
196 |
902 |
499.2 |
55.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.849 ± 0.571 | 2.965 ± 0.668 |
6.13 ± 0.718 | 5.369 ± 0.517 |
4.728 ± 0.579 | 6.771 ± 0.801 |
2.845 ± 0.338 | 4.327 ± 0.473 |
7.011 ± 0.765 | 10.617 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.963 ± 0.267 | 4.607 ± 0.126 |
5.769 ± 0.605 | 4.207 ± 0.226 |
4.006 ± 0.415 | 8.534 ± 0.695 |
5.529 ± 0.446 | 4.728 ± 1.06 |
1.683 ± 0.216 | 2.364 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
