
Umboniibacter marinipuniceus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Umboniibacter
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
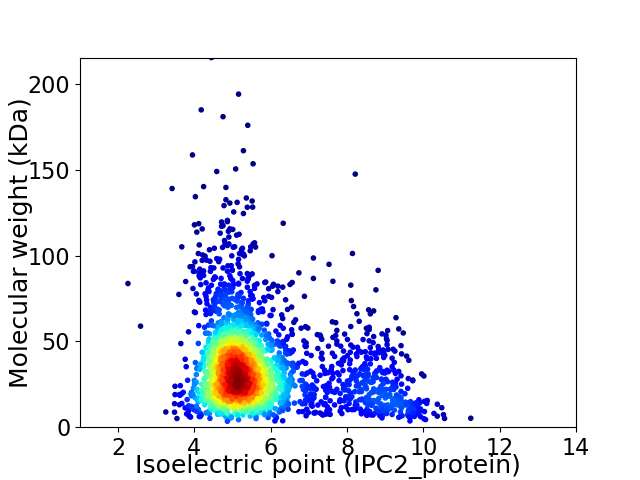
Virtual 2D-PAGE plot for 2413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M0A976|A0A3M0A976_9GAMM Glycerate dehydrogenase OS=Umboniibacter marinipuniceus OX=569599 GN=DFR27_1405 PE=3 SV=1
MM1 pKa = 7.34EE2 pKa = 5.86RR3 pKa = 11.84SLDD6 pKa = 3.56SRR8 pKa = 11.84LVIVSALLALTMSAVTSAQDD28 pKa = 3.7SEE30 pKa = 4.65IEE32 pKa = 4.23SSAVAAQSEE41 pKa = 5.08STTEE45 pKa = 4.05TEE47 pKa = 4.26AEE49 pKa = 4.25LPAVTYY55 pKa = 9.81EE56 pKa = 4.41SYY58 pKa = 11.2GSDD61 pKa = 2.82VGFVQTQPVPYY72 pKa = 10.22DD73 pKa = 3.57GDD75 pKa = 3.66PFEE78 pKa = 5.06DD79 pKa = 3.9LNRR82 pKa = 11.84AMLTFNDD89 pKa = 3.32VADD92 pKa = 5.14RR93 pKa = 11.84YY94 pKa = 10.54LLLPIVTGYY103 pKa = 11.05KK104 pKa = 10.39YY105 pKa = 9.27ITPDD109 pKa = 3.23PVEE112 pKa = 4.34RR113 pKa = 11.84SVDD116 pKa = 3.65NIFANLDD123 pKa = 4.28DD124 pKa = 4.86IGSAANSILQLKK136 pKa = 9.88IGDD139 pKa = 3.74AGVYY143 pKa = 10.03AGRR146 pKa = 11.84FLTNTTIGVLGIWDD160 pKa = 3.78VASEE164 pKa = 3.98LGMRR168 pKa = 11.84RR169 pKa = 11.84LEE171 pKa = 4.52GEE173 pKa = 4.75DD174 pKa = 3.34FGQTLGYY181 pKa = 10.08YY182 pKa = 9.04GVPEE186 pKa = 4.73GPYY189 pKa = 11.02LMLPFYY195 pKa = 10.87GPSTLRR201 pKa = 11.84DD202 pKa = 3.21APARR206 pKa = 11.84YY207 pKa = 9.15VDD209 pKa = 3.73SFVDD213 pKa = 3.42YY214 pKa = 10.13TSYY217 pKa = 11.21VDD219 pKa = 4.7HH220 pKa = 6.7VPTRR224 pKa = 11.84NSLMGVEE231 pKa = 4.21AVNVRR236 pKa = 11.84SQLIQAEE243 pKa = 4.16AFITGDD249 pKa = 3.02RR250 pKa = 11.84YY251 pKa = 9.04TFIRR255 pKa = 11.84DD256 pKa = 2.95AYY258 pKa = 8.84LQRR261 pKa = 11.84RR262 pKa = 11.84QYY264 pKa = 11.26LVLDD268 pKa = 3.83GQMPDD273 pKa = 4.27DD274 pKa = 4.28EE275 pKa = 5.06EE276 pKa = 5.16FDD278 pKa = 3.66EE279 pKa = 5.51FGGFGSAGEE288 pKa = 4.42SYY290 pKa = 10.86GGEE293 pKa = 4.26SYY295 pKa = 11.02GGEE298 pKa = 4.3SYY300 pKa = 11.02GGEE303 pKa = 4.3SYY305 pKa = 11.02GGEE308 pKa = 4.3SYY310 pKa = 11.02GGEE313 pKa = 4.3SYY315 pKa = 11.02GGEE318 pKa = 4.17SYY320 pKa = 11.25GGDD323 pKa = 3.41EE324 pKa = 5.42DD325 pKa = 5.59SDD327 pKa = 4.19PNRR330 pKa = 11.84DD331 pKa = 3.81NLSGSADD338 pKa = 3.81DD339 pKa = 6.01QIDD342 pKa = 3.87PPLNEE347 pKa = 3.72TDD349 pKa = 3.47GRR351 pKa = 11.84SDD353 pKa = 4.17NDD355 pKa = 3.74DD356 pKa = 4.45KK357 pKa = 11.65LPQDD361 pKa = 3.84SEE363 pKa = 4.47FF364 pKa = 3.99
MM1 pKa = 7.34EE2 pKa = 5.86RR3 pKa = 11.84SLDD6 pKa = 3.56SRR8 pKa = 11.84LVIVSALLALTMSAVTSAQDD28 pKa = 3.7SEE30 pKa = 4.65IEE32 pKa = 4.23SSAVAAQSEE41 pKa = 5.08STTEE45 pKa = 4.05TEE47 pKa = 4.26AEE49 pKa = 4.25LPAVTYY55 pKa = 9.81EE56 pKa = 4.41SYY58 pKa = 11.2GSDD61 pKa = 2.82VGFVQTQPVPYY72 pKa = 10.22DD73 pKa = 3.57GDD75 pKa = 3.66PFEE78 pKa = 5.06DD79 pKa = 3.9LNRR82 pKa = 11.84AMLTFNDD89 pKa = 3.32VADD92 pKa = 5.14RR93 pKa = 11.84YY94 pKa = 10.54LLLPIVTGYY103 pKa = 11.05KK104 pKa = 10.39YY105 pKa = 9.27ITPDD109 pKa = 3.23PVEE112 pKa = 4.34RR113 pKa = 11.84SVDD116 pKa = 3.65NIFANLDD123 pKa = 4.28DD124 pKa = 4.86IGSAANSILQLKK136 pKa = 9.88IGDD139 pKa = 3.74AGVYY143 pKa = 10.03AGRR146 pKa = 11.84FLTNTTIGVLGIWDD160 pKa = 3.78VASEE164 pKa = 3.98LGMRR168 pKa = 11.84RR169 pKa = 11.84LEE171 pKa = 4.52GEE173 pKa = 4.75DD174 pKa = 3.34FGQTLGYY181 pKa = 10.08YY182 pKa = 9.04GVPEE186 pKa = 4.73GPYY189 pKa = 11.02LMLPFYY195 pKa = 10.87GPSTLRR201 pKa = 11.84DD202 pKa = 3.21APARR206 pKa = 11.84YY207 pKa = 9.15VDD209 pKa = 3.73SFVDD213 pKa = 3.42YY214 pKa = 10.13TSYY217 pKa = 11.21VDD219 pKa = 4.7HH220 pKa = 6.7VPTRR224 pKa = 11.84NSLMGVEE231 pKa = 4.21AVNVRR236 pKa = 11.84SQLIQAEE243 pKa = 4.16AFITGDD249 pKa = 3.02RR250 pKa = 11.84YY251 pKa = 9.04TFIRR255 pKa = 11.84DD256 pKa = 2.95AYY258 pKa = 8.84LQRR261 pKa = 11.84RR262 pKa = 11.84QYY264 pKa = 11.26LVLDD268 pKa = 3.83GQMPDD273 pKa = 4.27DD274 pKa = 4.28EE275 pKa = 5.06EE276 pKa = 5.16FDD278 pKa = 3.66EE279 pKa = 5.51FGGFGSAGEE288 pKa = 4.42SYY290 pKa = 10.86GGEE293 pKa = 4.26SYY295 pKa = 11.02GGEE298 pKa = 4.3SYY300 pKa = 11.02GGEE303 pKa = 4.3SYY305 pKa = 11.02GGEE308 pKa = 4.3SYY310 pKa = 11.02GGEE313 pKa = 4.3SYY315 pKa = 11.02GGEE318 pKa = 4.17SYY320 pKa = 11.25GGDD323 pKa = 3.41EE324 pKa = 5.42DD325 pKa = 5.59SDD327 pKa = 4.19PNRR330 pKa = 11.84DD331 pKa = 3.81NLSGSADD338 pKa = 3.81DD339 pKa = 6.01QIDD342 pKa = 3.87PPLNEE347 pKa = 3.72TDD349 pKa = 3.47GRR351 pKa = 11.84SDD353 pKa = 4.17NDD355 pKa = 3.74DD356 pKa = 4.45KK357 pKa = 11.65LPQDD361 pKa = 3.84SEE363 pKa = 4.47FF364 pKa = 3.99
Molecular weight: 39.62 kDa
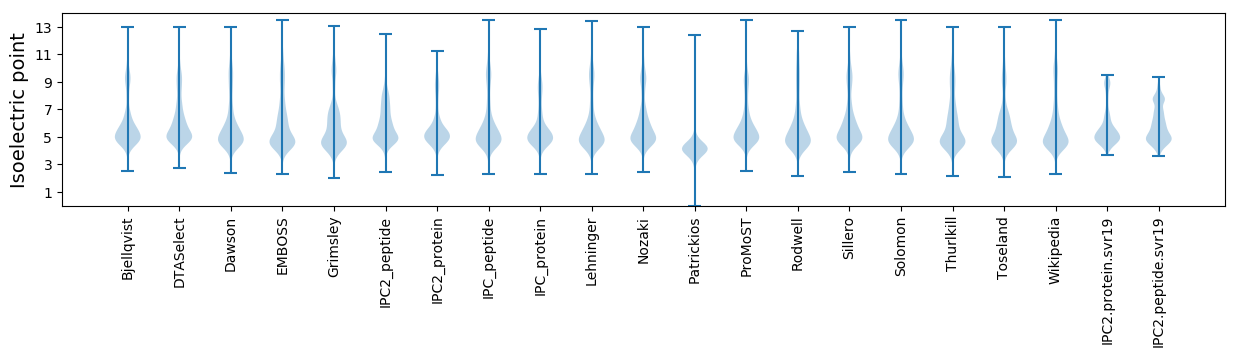
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M0AB83|A0A3M0AB83_9GAMM Glycine dehydrogenase (decarboxylating) OS=Umboniibacter marinipuniceus OX=569599 GN=gcvP PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.39GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.39GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
804001 |
29 |
1961 |
333.2 |
36.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.053 ± 0.054 | 0.969 ± 0.014 |
5.779 ± 0.045 | 6.437 ± 0.05 |
3.986 ± 0.031 | 7.251 ± 0.044 |
2.128 ± 0.023 | 5.822 ± 0.037 |
3.818 ± 0.039 | 10.335 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.369 ± 0.022 | 3.81 ± 0.027 |
4.023 ± 0.026 | 4.179 ± 0.031 |
5.383 ± 0.041 | 6.915 ± 0.043 |
5.205 ± 0.037 | 7.331 ± 0.037 |
1.337 ± 0.022 | 2.869 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
