
Arthrobacter nitrophenolicus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
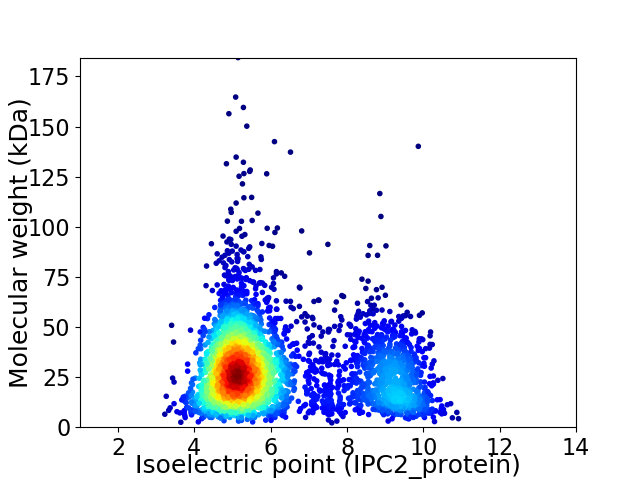
Virtual 2D-PAGE plot for 3633 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
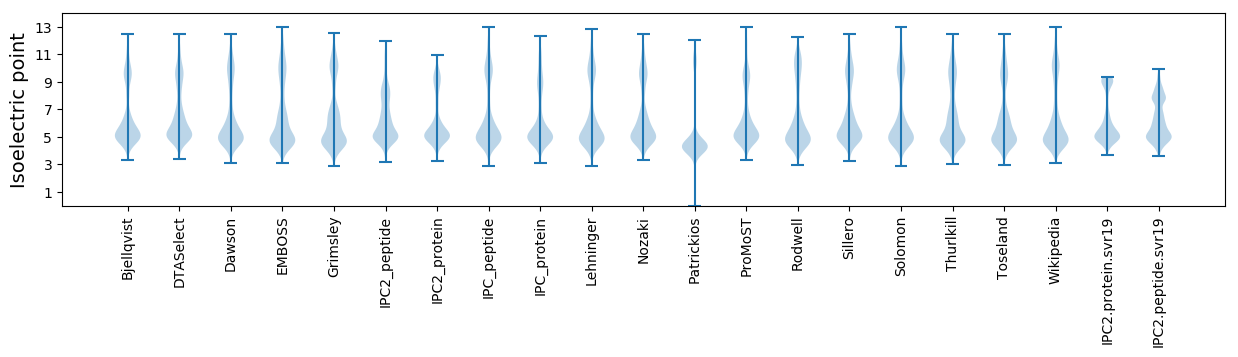
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8TT60|L8TT60_9MICC DUF948 domain-containing protein OS=Arthrobacter nitrophenolicus OX=683150 GN=E2R57_05835 PE=4 SV=1
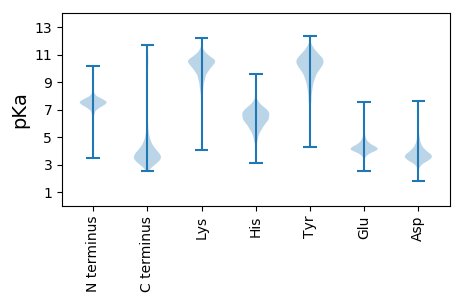
MM1 pKa = 6.85LTAYY5 pKa = 10.23YY6 pKa = 10.84VLLDD10 pKa = 4.5DD11 pKa = 5.31GGSSGVRR18 pKa = 11.84FGCNDD23 pKa = 3.41SLVGLARR30 pKa = 11.84AAPGTEE36 pKa = 4.04PLLTAAMNALLQGAGDD52 pKa = 4.17ADD54 pKa = 3.78IPLDD58 pKa = 4.57PIEE61 pKa = 4.67VSDD64 pKa = 4.98VYY66 pKa = 11.26NALDD70 pKa = 3.81GSEE73 pKa = 4.22LTFLSGSFDD82 pKa = 3.37GTTVTVYY89 pKa = 10.84LAGTLSLGGACDD101 pKa = 3.67VPRR104 pKa = 11.84VEE106 pKa = 5.22AQLTHH111 pKa = 5.84TAVAAVGAIRR121 pKa = 11.84ADD123 pKa = 3.7VYY125 pKa = 11.87VNGQPLSEE133 pKa = 4.58VLRR136 pKa = 11.84QDD138 pKa = 2.98
MM1 pKa = 6.85LTAYY5 pKa = 10.23YY6 pKa = 10.84VLLDD10 pKa = 4.5DD11 pKa = 5.31GGSSGVRR18 pKa = 11.84FGCNDD23 pKa = 3.41SLVGLARR30 pKa = 11.84AAPGTEE36 pKa = 4.04PLLTAAMNALLQGAGDD52 pKa = 4.17ADD54 pKa = 3.78IPLDD58 pKa = 4.57PIEE61 pKa = 4.67VSDD64 pKa = 4.98VYY66 pKa = 11.26NALDD70 pKa = 3.81GSEE73 pKa = 4.22LTFLSGSFDD82 pKa = 3.37GTTVTVYY89 pKa = 10.84LAGTLSLGGACDD101 pKa = 3.67VPRR104 pKa = 11.84VEE106 pKa = 5.22AQLTHH111 pKa = 5.84TAVAAVGAIRR121 pKa = 11.84ADD123 pKa = 3.7VYY125 pKa = 11.87VNGQPLSEE133 pKa = 4.58VLRR136 pKa = 11.84QDD138 pKa = 2.98
Molecular weight: 14.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8TU56|L8TU56_9MICC Extracellular solute-binding protein family 1 protein OS=Arthrobacter nitrophenolicus OX=683150 GN=G205_09558 PE=4 SV=1
MM1 pKa = 7.64SSDD4 pKa = 4.34ALVADD9 pKa = 4.2QTDD12 pKa = 3.75HH13 pKa = 6.84PGEE16 pKa = 4.26PVAVTAAGALPWRR29 pKa = 11.84VTNDD33 pKa = 3.19RR34 pKa = 11.84LEE36 pKa = 3.92VLLIHH41 pKa = 7.32RR42 pKa = 11.84PRR44 pKa = 11.84YY45 pKa = 9.88DD46 pKa = 3.09DD47 pKa = 3.55WSWPKK52 pKa = 11.01GKK54 pKa = 9.5IDD56 pKa = 4.39DD57 pKa = 4.67GEE59 pKa = 4.58TIPEE63 pKa = 4.04CALRR67 pKa = 11.84EE68 pKa = 3.99VRR70 pKa = 11.84EE71 pKa = 4.3EE72 pKa = 4.02IGLDD76 pKa = 3.39APLGRR81 pKa = 11.84PLPAIHH87 pKa = 6.16YY88 pKa = 8.76RR89 pKa = 11.84VASGLKK95 pKa = 9.62VVFYY99 pKa = 9.98WAVKK103 pKa = 10.22ANGTRR108 pKa = 11.84LVPDD112 pKa = 3.97GKK114 pKa = 10.86EE115 pKa = 3.7VDD117 pKa = 3.81SVMWCAPEE125 pKa = 4.02KK126 pKa = 10.36AAKK129 pKa = 10.14LLSNPTDD136 pKa = 3.78VVPLEE141 pKa = 4.31YY142 pKa = 10.23LRR144 pKa = 11.84KK145 pKa = 8.91AHH147 pKa = 7.16EE148 pKa = 4.42EE149 pKa = 4.49GEE151 pKa = 4.45LNTWPLVSSGMPKK164 pKa = 9.71RR165 pKa = 11.84SHH167 pKa = 6.49ARR169 pKa = 11.84PGPKK173 pKa = 9.55PKK175 pKa = 10.57VNARR179 pKa = 11.84WPPPAPGRR187 pKa = 11.84HH188 pKa = 5.22RR189 pKa = 11.84RR190 pKa = 11.84WDD192 pKa = 3.38GCCRR196 pKa = 11.84YY197 pKa = 9.91GSPRR201 pKa = 11.84AWSPVPGSGAWRR213 pKa = 11.84LLHH216 pKa = 7.2PMRR219 pKa = 11.84KK220 pKa = 7.89WQAQRR225 pKa = 11.84FGWRR229 pKa = 11.84KK230 pKa = 8.67PSRR233 pKa = 11.84NTATTAARR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84PRR245 pKa = 11.84PSSRR249 pKa = 11.84PSSISSGRR257 pKa = 11.84WWSAPTAPRR266 pKa = 11.84CRR268 pKa = 11.84RR269 pKa = 11.84SSRR272 pKa = 11.84SWRR275 pKa = 11.84STCPAGWAACCPRR288 pKa = 11.84RR289 pKa = 11.84SHH291 pKa = 7.32ISPPANWWSAMWRR304 pKa = 11.84PGQRR308 pKa = 11.84TGWSPSSSSSRR319 pKa = 11.84STTDD323 pKa = 3.12PAGIQEE329 pKa = 4.44ADD331 pKa = 3.31KK332 pKa = 11.09QPRR335 pKa = 3.27
MM1 pKa = 7.64SSDD4 pKa = 4.34ALVADD9 pKa = 4.2QTDD12 pKa = 3.75HH13 pKa = 6.84PGEE16 pKa = 4.26PVAVTAAGALPWRR29 pKa = 11.84VTNDD33 pKa = 3.19RR34 pKa = 11.84LEE36 pKa = 3.92VLLIHH41 pKa = 7.32RR42 pKa = 11.84PRR44 pKa = 11.84YY45 pKa = 9.88DD46 pKa = 3.09DD47 pKa = 3.55WSWPKK52 pKa = 11.01GKK54 pKa = 9.5IDD56 pKa = 4.39DD57 pKa = 4.67GEE59 pKa = 4.58TIPEE63 pKa = 4.04CALRR67 pKa = 11.84EE68 pKa = 3.99VRR70 pKa = 11.84EE71 pKa = 4.3EE72 pKa = 4.02IGLDD76 pKa = 3.39APLGRR81 pKa = 11.84PLPAIHH87 pKa = 6.16YY88 pKa = 8.76RR89 pKa = 11.84VASGLKK95 pKa = 9.62VVFYY99 pKa = 9.98WAVKK103 pKa = 10.22ANGTRR108 pKa = 11.84LVPDD112 pKa = 3.97GKK114 pKa = 10.86EE115 pKa = 3.7VDD117 pKa = 3.81SVMWCAPEE125 pKa = 4.02KK126 pKa = 10.36AAKK129 pKa = 10.14LLSNPTDD136 pKa = 3.78VVPLEE141 pKa = 4.31YY142 pKa = 10.23LRR144 pKa = 11.84KK145 pKa = 8.91AHH147 pKa = 7.16EE148 pKa = 4.42EE149 pKa = 4.49GEE151 pKa = 4.45LNTWPLVSSGMPKK164 pKa = 9.71RR165 pKa = 11.84SHH167 pKa = 6.49ARR169 pKa = 11.84PGPKK173 pKa = 9.55PKK175 pKa = 10.57VNARR179 pKa = 11.84WPPPAPGRR187 pKa = 11.84HH188 pKa = 5.22RR189 pKa = 11.84RR190 pKa = 11.84WDD192 pKa = 3.38GCCRR196 pKa = 11.84YY197 pKa = 9.91GSPRR201 pKa = 11.84AWSPVPGSGAWRR213 pKa = 11.84LLHH216 pKa = 7.2PMRR219 pKa = 11.84KK220 pKa = 7.89WQAQRR225 pKa = 11.84FGWRR229 pKa = 11.84KK230 pKa = 8.67PSRR233 pKa = 11.84NTATTAARR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84PRR245 pKa = 11.84PSSRR249 pKa = 11.84PSSISSGRR257 pKa = 11.84WWSAPTAPRR266 pKa = 11.84CRR268 pKa = 11.84RR269 pKa = 11.84SSRR272 pKa = 11.84SWRR275 pKa = 11.84STCPAGWAACCPRR288 pKa = 11.84RR289 pKa = 11.84SHH291 pKa = 7.32ISPPANWWSAMWRR304 pKa = 11.84PGQRR308 pKa = 11.84TGWSPSSSSSRR319 pKa = 11.84STTDD323 pKa = 3.12PAGIQEE329 pKa = 4.44ADD331 pKa = 3.31KK332 pKa = 11.09QPRR335 pKa = 3.27
Molecular weight: 37.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1041394 |
20 |
1758 |
286.6 |
30.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.219 ± 0.058 | 0.652 ± 0.011 |
5.587 ± 0.034 | 5.651 ± 0.04 |
3.2 ± 0.027 | 9.163 ± 0.036 |
2.114 ± 0.021 | 4.197 ± 0.027 |
2.719 ± 0.03 | 10.242 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.997 ± 0.017 | 2.427 ± 0.023 |
5.632 ± 0.04 | 3.194 ± 0.022 |
6.619 ± 0.043 | 5.783 ± 0.03 |
5.796 ± 0.033 | 8.268 ± 0.036 |
1.47 ± 0.018 | 2.071 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
