
Wallemia hederae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Wallemiomycotina; Wallemiomycetes; Wallemiales; Wallemiaceae; Wallemia
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
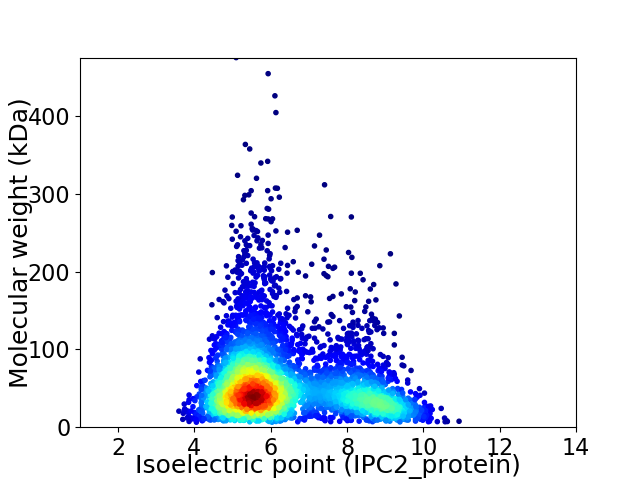
Virtual 2D-PAGE plot for 4107 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4T0FCD5|A0A4T0FCD5_9BASI Aminotran_1_2 domain-containing protein OS=Wallemia hederae OX=1540922 GN=E3P99_03898 PE=4 SV=1
MM1 pKa = 7.03VGKK4 pKa = 10.51FGLLSLAAVAGVAAAQSKK22 pKa = 10.17DD23 pKa = 3.49YY24 pKa = 10.82PGVVSANDD32 pKa = 3.59NGPTNPDD39 pKa = 3.2QPEE42 pKa = 4.58LGTDD46 pKa = 3.41TPEE49 pKa = 4.47DD50 pKa = 3.61SEE52 pKa = 5.14ARR54 pKa = 11.84LLTVNNIDD62 pKa = 4.06DD63 pKa = 3.92FCIFGPKK70 pKa = 10.09DD71 pKa = 3.2ADD73 pKa = 3.46KK74 pKa = 10.98TIGEE78 pKa = 4.44TEE80 pKa = 4.03AEE82 pKa = 4.08QVAWCTQPRR91 pKa = 11.84NNARR95 pKa = 11.84LIPEE99 pKa = 3.97DD100 pKa = 4.19TFTGLHH106 pKa = 5.54FVKK109 pKa = 9.94TPYY112 pKa = 10.39YY113 pKa = 9.96IQLQGWGDD121 pKa = 3.56LTKK124 pKa = 10.45IGIKK128 pKa = 10.61DD129 pKa = 3.23GDD131 pKa = 3.9YY132 pKa = 11.17GGEE135 pKa = 4.14LDD137 pKa = 3.57PHH139 pKa = 5.94GAKK142 pKa = 10.57DD143 pKa = 3.69LGNPIGGNVTSNISGEE159 pKa = 3.99ATFYY163 pKa = 10.39EE164 pKa = 4.5EE165 pKa = 3.5WMEE168 pKa = 4.49FISYY172 pKa = 9.4EE173 pKa = 4.14QFCLRR178 pKa = 11.84VCTAGDD184 pKa = 4.06DD185 pKa = 3.68TWDD188 pKa = 3.44AATMCEE194 pKa = 4.08HH195 pKa = 7.14KK196 pKa = 10.25LDD198 pKa = 3.76EE199 pKa = 4.6MGCGFIMPGDD209 pKa = 3.85YY210 pKa = 9.93QGKK213 pKa = 8.18QDD215 pKa = 4.48QFVSCDD221 pKa = 3.08ADD223 pKa = 3.43AAFPPGVYY231 pKa = 9.46PDD233 pKa = 3.71GDD235 pKa = 3.83GYY237 pKa = 10.24STFNQRR243 pKa = 11.84YY244 pKa = 6.64TGTLTSNTYY253 pKa = 9.38EE254 pKa = 4.21VYY256 pKa = 10.05TVGDD260 pKa = 3.78TATPSGPYY268 pKa = 8.6STPSSSNCKK277 pKa = 8.08TQDD280 pKa = 3.37GLNTYY285 pKa = 10.28VNTDD289 pKa = 3.22APQSTGSSDD298 pKa = 3.38NNTNAEE304 pKa = 4.25NGDD307 pKa = 3.81SGAGSVALGAVATVSIALISAFAILSS333 pKa = 3.64
MM1 pKa = 7.03VGKK4 pKa = 10.51FGLLSLAAVAGVAAAQSKK22 pKa = 10.17DD23 pKa = 3.49YY24 pKa = 10.82PGVVSANDD32 pKa = 3.59NGPTNPDD39 pKa = 3.2QPEE42 pKa = 4.58LGTDD46 pKa = 3.41TPEE49 pKa = 4.47DD50 pKa = 3.61SEE52 pKa = 5.14ARR54 pKa = 11.84LLTVNNIDD62 pKa = 4.06DD63 pKa = 3.92FCIFGPKK70 pKa = 10.09DD71 pKa = 3.2ADD73 pKa = 3.46KK74 pKa = 10.98TIGEE78 pKa = 4.44TEE80 pKa = 4.03AEE82 pKa = 4.08QVAWCTQPRR91 pKa = 11.84NNARR95 pKa = 11.84LIPEE99 pKa = 3.97DD100 pKa = 4.19TFTGLHH106 pKa = 5.54FVKK109 pKa = 9.94TPYY112 pKa = 10.39YY113 pKa = 9.96IQLQGWGDD121 pKa = 3.56LTKK124 pKa = 10.45IGIKK128 pKa = 10.61DD129 pKa = 3.23GDD131 pKa = 3.9YY132 pKa = 11.17GGEE135 pKa = 4.14LDD137 pKa = 3.57PHH139 pKa = 5.94GAKK142 pKa = 10.57DD143 pKa = 3.69LGNPIGGNVTSNISGEE159 pKa = 3.99ATFYY163 pKa = 10.39EE164 pKa = 4.5EE165 pKa = 3.5WMEE168 pKa = 4.49FISYY172 pKa = 9.4EE173 pKa = 4.14QFCLRR178 pKa = 11.84VCTAGDD184 pKa = 4.06DD185 pKa = 3.68TWDD188 pKa = 3.44AATMCEE194 pKa = 4.08HH195 pKa = 7.14KK196 pKa = 10.25LDD198 pKa = 3.76EE199 pKa = 4.6MGCGFIMPGDD209 pKa = 3.85YY210 pKa = 9.93QGKK213 pKa = 8.18QDD215 pKa = 4.48QFVSCDD221 pKa = 3.08ADD223 pKa = 3.43AAFPPGVYY231 pKa = 9.46PDD233 pKa = 3.71GDD235 pKa = 3.83GYY237 pKa = 10.24STFNQRR243 pKa = 11.84YY244 pKa = 6.64TGTLTSNTYY253 pKa = 9.38EE254 pKa = 4.21VYY256 pKa = 10.05TVGDD260 pKa = 3.78TATPSGPYY268 pKa = 8.6STPSSSNCKK277 pKa = 8.08TQDD280 pKa = 3.37GLNTYY285 pKa = 10.28VNTDD289 pKa = 3.22APQSTGSSDD298 pKa = 3.38NNTNAEE304 pKa = 4.25NGDD307 pKa = 3.81SGAGSVALGAVATVSIALISAFAILSS333 pKa = 3.64
Molecular weight: 35.27 kDa
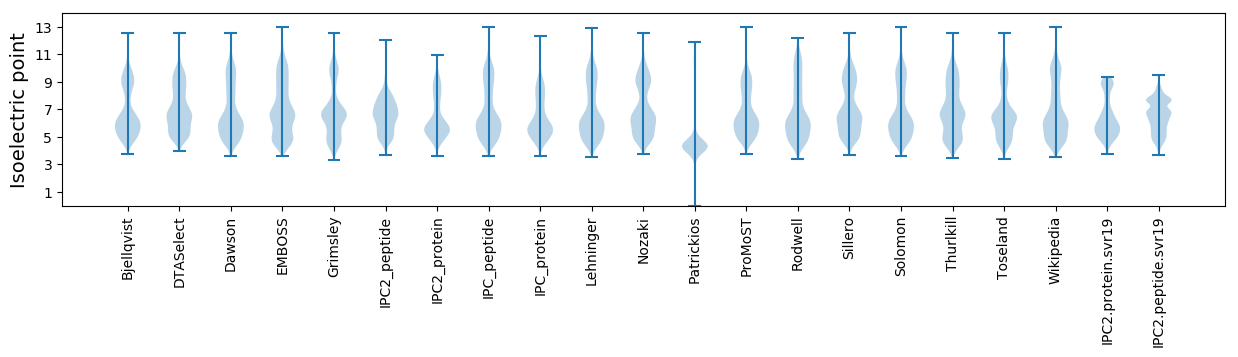
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4T0FKS1|A0A4T0FKS1_9BASI Isoleucyl-tRNA synthetase OS=Wallemia hederae OX=1540922 GN=E3P99_02380 PE=3 SV=1
MM1 pKa = 7.82ASPATVFSPTLNALNQLRR19 pKa = 11.84FASRR23 pKa = 11.84GTDD26 pKa = 3.54YY27 pKa = 11.19QPSTRR32 pKa = 11.84KK33 pKa = 9.59RR34 pKa = 11.84KK35 pKa = 8.51RR36 pKa = 11.84TFGFLARR43 pKa = 11.84IRR45 pKa = 11.84SRR47 pKa = 11.84TGRR50 pKa = 11.84KK51 pKa = 9.05IIARR55 pKa = 11.84RR56 pKa = 11.84LKK58 pKa = 10.34RR59 pKa = 11.84GRR61 pKa = 11.84KK62 pKa = 8.89NMSHH66 pKa = 6.79
MM1 pKa = 7.82ASPATVFSPTLNALNQLRR19 pKa = 11.84FASRR23 pKa = 11.84GTDD26 pKa = 3.54YY27 pKa = 11.19QPSTRR32 pKa = 11.84KK33 pKa = 9.59RR34 pKa = 11.84KK35 pKa = 8.51RR36 pKa = 11.84TFGFLARR43 pKa = 11.84IRR45 pKa = 11.84SRR47 pKa = 11.84TGRR50 pKa = 11.84KK51 pKa = 9.05IIARR55 pKa = 11.84RR56 pKa = 11.84LKK58 pKa = 10.34RR59 pKa = 11.84GRR61 pKa = 11.84KK62 pKa = 8.89NMSHH66 pKa = 6.79
Molecular weight: 7.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2297997 |
59 |
4344 |
559.5 |
62.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.12 ± 0.031 | 1.029 ± 0.012 |
6.308 ± 0.027 | 6.179 ± 0.034 |
3.578 ± 0.022 | 5.863 ± 0.031 |
2.597 ± 0.017 | 5.157 ± 0.024 |
5.862 ± 0.031 | 9.168 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.012 | 4.381 ± 0.02 |
5.143 ± 0.032 | 4.548 ± 0.034 |
5.422 ± 0.025 | 8.784 ± 0.038 |
5.748 ± 0.02 | 6.07 ± 0.028 |
1.101 ± 0.011 | 2.887 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
