
Streptococcus satellite phage Javan256
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
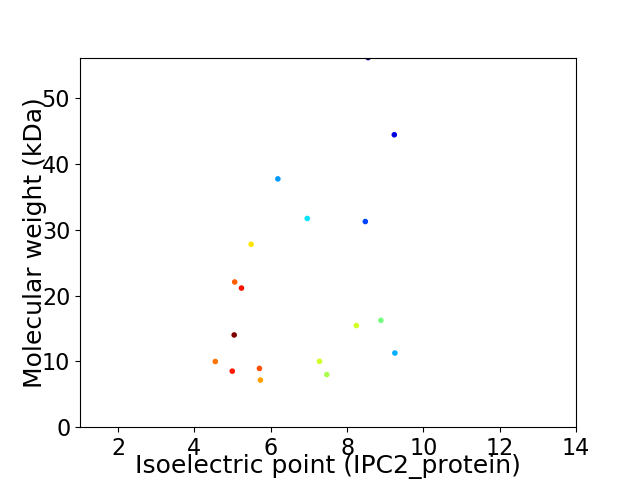
Virtual 2D-PAGE plot for 18 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZMJ1|A0A4D5ZMJ1_9VIRU RelB/StbD replicon stabilization protein OS=Streptococcus satellite phage Javan256 OX=2558593 GN=JavanS256_0016 PE=4 SV=1
MM1 pKa = 6.53TTEE4 pKa = 4.46QILDD8 pKa = 3.79SYY10 pKa = 11.58EE11 pKa = 4.31SLTSDD16 pKa = 2.9MMKK19 pKa = 10.43AVSALNIVRR28 pKa = 11.84RR29 pKa = 11.84WLEE32 pKa = 3.73NVKK35 pKa = 10.61KK36 pKa = 10.47SDD38 pKa = 2.86EE39 pKa = 4.28GYY41 pKa = 10.7YY42 pKa = 10.85DD43 pKa = 5.25FIAFKK48 pKa = 10.1EE49 pKa = 4.36AYY51 pKa = 10.13ADD53 pKa = 4.02IINLAMNDD61 pKa = 3.86LQRR64 pKa = 11.84LASEE68 pKa = 4.33HH69 pKa = 5.95YY70 pKa = 10.41KK71 pKa = 10.65FIEE74 pKa = 4.1RR75 pKa = 11.84AEE77 pKa = 3.95KK78 pKa = 11.0DD79 pKa = 3.51EE80 pKa = 4.43FSEE83 pKa = 5.22AEE85 pKa = 3.87
MM1 pKa = 6.53TTEE4 pKa = 4.46QILDD8 pKa = 3.79SYY10 pKa = 11.58EE11 pKa = 4.31SLTSDD16 pKa = 2.9MMKK19 pKa = 10.43AVSALNIVRR28 pKa = 11.84RR29 pKa = 11.84WLEE32 pKa = 3.73NVKK35 pKa = 10.61KK36 pKa = 10.47SDD38 pKa = 2.86EE39 pKa = 4.28GYY41 pKa = 10.7YY42 pKa = 10.85DD43 pKa = 5.25FIAFKK48 pKa = 10.1EE49 pKa = 4.36AYY51 pKa = 10.13ADD53 pKa = 4.02IINLAMNDD61 pKa = 3.86LQRR64 pKa = 11.84LASEE68 pKa = 4.33HH69 pKa = 5.95YY70 pKa = 10.41KK71 pKa = 10.65FIEE74 pKa = 4.1RR75 pKa = 11.84AEE77 pKa = 3.95KK78 pKa = 11.0DD79 pKa = 3.51EE80 pKa = 4.43FSEE83 pKa = 5.22AEE85 pKa = 3.87
Molecular weight: 9.98 kDa
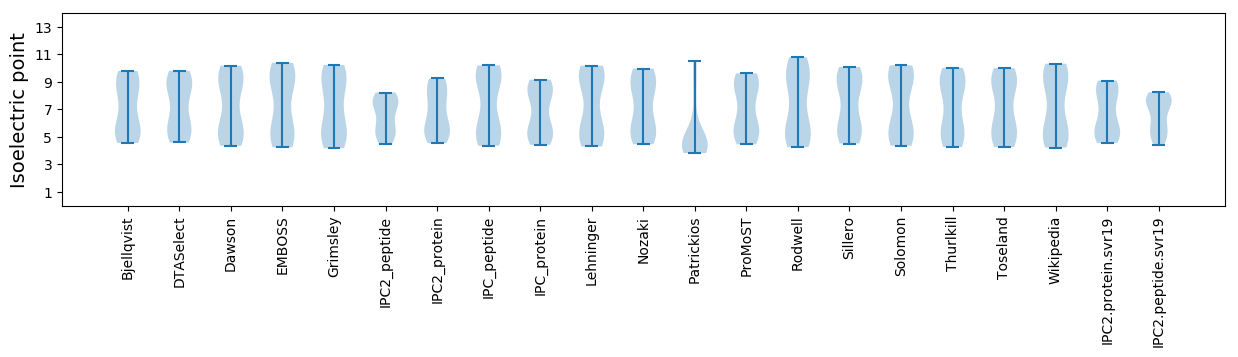
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZH88|A0A4D5ZH88_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan256 OX=2558593 GN=JavanS256_0011 PE=4 SV=1
MM1 pKa = 7.55NIKK4 pKa = 10.55DD5 pKa = 3.92KK6 pKa = 10.89IKK8 pKa = 11.03KK9 pKa = 9.53NGQKK13 pKa = 10.04VYY15 pKa = 10.21YY16 pKa = 9.67ASVYY20 pKa = 10.7LGVDD24 pKa = 2.95QLTGKK29 pKa = 10.14KK30 pKa = 10.31ARR32 pKa = 11.84TTVTATTKK40 pKa = 10.63KK41 pKa = 10.27GVKK44 pKa = 8.67TKK46 pKa = 10.83ARR48 pKa = 11.84DD49 pKa = 3.59VINAFAANGYY59 pKa = 5.7TVKK62 pKa = 10.68DD63 pKa = 3.48KK64 pKa = 9.74PTITTYY70 pKa = 11.08KK71 pKa = 10.18EE72 pKa = 4.03LVKK75 pKa = 10.72VWWDD79 pKa = 3.31SYY81 pKa = 11.64KK82 pKa = 10.2NTVKK86 pKa = 10.73PNTRR90 pKa = 11.84QSMEE94 pKa = 3.66GLVRR98 pKa = 11.84VHH100 pKa = 6.91LLPVFGDD107 pKa = 3.68YY108 pKa = 10.69KK109 pKa = 10.62LSKK112 pKa = 9.42LTTPIIQHH120 pKa = 5.63QVNKK124 pKa = 9.59WADD127 pKa = 3.39KK128 pKa = 11.42ANTEE132 pKa = 4.1QKK134 pKa = 10.97GAFTNYY140 pKa = 10.64SLLHH144 pKa = 5.79NMNKK148 pKa = 10.25RR149 pKa = 11.84ILKK152 pKa = 10.11YY153 pKa = 10.19GVSLQVIPHH162 pKa = 6.2NPANDD167 pKa = 3.43VIVPRR172 pKa = 11.84KK173 pKa = 8.24RR174 pKa = 11.84QKK176 pKa = 10.72EE177 pKa = 4.15KK178 pKa = 8.73TTVKK182 pKa = 10.72YY183 pKa = 10.72LGNKK187 pKa = 7.16EE188 pKa = 4.11LKK190 pKa = 10.43QFLDD194 pKa = 3.68YY195 pKa = 11.33LDD197 pKa = 4.79TLDD200 pKa = 3.68QSNYY204 pKa = 9.49EE205 pKa = 3.97NLFDD209 pKa = 3.81VVLYY213 pKa = 8.25KK214 pKa = 10.5TLLATGCRR222 pKa = 11.84ISEE225 pKa = 4.1ALALEE230 pKa = 4.39WSDD233 pKa = 3.55IDD235 pKa = 4.56LDD237 pKa = 4.06NGIISINKK245 pKa = 7.21TLNRR249 pKa = 11.84YY250 pKa = 7.65QEE252 pKa = 4.1INSPKK257 pKa = 10.13SSAGYY262 pKa = 10.09RR263 pKa = 11.84DD264 pKa = 3.43IPIDD268 pKa = 3.67NATLLMLKK276 pKa = 9.77QYY278 pKa = 11.18KK279 pKa = 9.65NRR281 pKa = 11.84QQVQSWQLGRR291 pKa = 11.84SEE293 pKa = 4.28TVVFSVFTEE302 pKa = 4.94KK303 pKa = 10.81YY304 pKa = 10.17AYY306 pKa = 10.08SCNLRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84LDD315 pKa = 3.44KK316 pKa = 10.72HH317 pKa = 6.28FKK319 pKa = 9.95AAGVTNVSFHH329 pKa = 6.03GFRR332 pKa = 11.84HH333 pKa = 4.73THH335 pKa = 4.12TTMMLYY341 pKa = 10.52AQASPKK347 pKa = 10.17DD348 pKa = 3.49VQYY351 pKa = 11.49RR352 pKa = 11.84LGHH355 pKa = 6.09SSLMMTEE362 pKa = 3.5NVYY365 pKa = 9.01WHH367 pKa = 6.72TNQEE371 pKa = 4.08NAKK374 pKa = 9.53KK375 pKa = 10.29AVSNYY380 pKa = 6.06EE381 pKa = 3.8TAINSLL387 pKa = 3.69
MM1 pKa = 7.55NIKK4 pKa = 10.55DD5 pKa = 3.92KK6 pKa = 10.89IKK8 pKa = 11.03KK9 pKa = 9.53NGQKK13 pKa = 10.04VYY15 pKa = 10.21YY16 pKa = 9.67ASVYY20 pKa = 10.7LGVDD24 pKa = 2.95QLTGKK29 pKa = 10.14KK30 pKa = 10.31ARR32 pKa = 11.84TTVTATTKK40 pKa = 10.63KK41 pKa = 10.27GVKK44 pKa = 8.67TKK46 pKa = 10.83ARR48 pKa = 11.84DD49 pKa = 3.59VINAFAANGYY59 pKa = 5.7TVKK62 pKa = 10.68DD63 pKa = 3.48KK64 pKa = 9.74PTITTYY70 pKa = 11.08KK71 pKa = 10.18EE72 pKa = 4.03LVKK75 pKa = 10.72VWWDD79 pKa = 3.31SYY81 pKa = 11.64KK82 pKa = 10.2NTVKK86 pKa = 10.73PNTRR90 pKa = 11.84QSMEE94 pKa = 3.66GLVRR98 pKa = 11.84VHH100 pKa = 6.91LLPVFGDD107 pKa = 3.68YY108 pKa = 10.69KK109 pKa = 10.62LSKK112 pKa = 9.42LTTPIIQHH120 pKa = 5.63QVNKK124 pKa = 9.59WADD127 pKa = 3.39KK128 pKa = 11.42ANTEE132 pKa = 4.1QKK134 pKa = 10.97GAFTNYY140 pKa = 10.64SLLHH144 pKa = 5.79NMNKK148 pKa = 10.25RR149 pKa = 11.84ILKK152 pKa = 10.11YY153 pKa = 10.19GVSLQVIPHH162 pKa = 6.2NPANDD167 pKa = 3.43VIVPRR172 pKa = 11.84KK173 pKa = 8.24RR174 pKa = 11.84QKK176 pKa = 10.72EE177 pKa = 4.15KK178 pKa = 8.73TTVKK182 pKa = 10.72YY183 pKa = 10.72LGNKK187 pKa = 7.16EE188 pKa = 4.11LKK190 pKa = 10.43QFLDD194 pKa = 3.68YY195 pKa = 11.33LDD197 pKa = 4.79TLDD200 pKa = 3.68QSNYY204 pKa = 9.49EE205 pKa = 3.97NLFDD209 pKa = 3.81VVLYY213 pKa = 8.25KK214 pKa = 10.5TLLATGCRR222 pKa = 11.84ISEE225 pKa = 4.1ALALEE230 pKa = 4.39WSDD233 pKa = 3.55IDD235 pKa = 4.56LDD237 pKa = 4.06NGIISINKK245 pKa = 7.21TLNRR249 pKa = 11.84YY250 pKa = 7.65QEE252 pKa = 4.1INSPKK257 pKa = 10.13SSAGYY262 pKa = 10.09RR263 pKa = 11.84DD264 pKa = 3.43IPIDD268 pKa = 3.67NATLLMLKK276 pKa = 9.77QYY278 pKa = 11.18KK279 pKa = 9.65NRR281 pKa = 11.84QQVQSWQLGRR291 pKa = 11.84SEE293 pKa = 4.28TVVFSVFTEE302 pKa = 4.94KK303 pKa = 10.81YY304 pKa = 10.17AYY306 pKa = 10.08SCNLRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84LDD315 pKa = 3.44KK316 pKa = 10.72HH317 pKa = 6.28FKK319 pKa = 9.95AAGVTNVSFHH329 pKa = 6.03GFRR332 pKa = 11.84HH333 pKa = 4.73THH335 pKa = 4.12TTMMLYY341 pKa = 10.52AQASPKK347 pKa = 10.17DD348 pKa = 3.49VQYY351 pKa = 11.49RR352 pKa = 11.84LGHH355 pKa = 6.09SSLMMTEE362 pKa = 3.5NVYY365 pKa = 9.01WHH367 pKa = 6.72TNQEE371 pKa = 4.08NAKK374 pKa = 9.53KK375 pKa = 10.29AVSNYY380 pKa = 6.06EE381 pKa = 3.8TAINSLL387 pKa = 3.69
Molecular weight: 44.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3301 |
62 |
483 |
183.4 |
21.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.119 ± 0.413 | 0.636 ± 0.213 |
5.332 ± 0.254 | 7.907 ± 0.626 |
3.666 ± 0.413 | 4.605 ± 0.297 |
2.181 ± 0.304 | 6.513 ± 0.716 |
9.391 ± 0.432 | 9.603 ± 0.365 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.193 | 5.029 ± 0.472 |
2.939 ± 0.388 | 4.968 ± 0.261 |
4.756 ± 0.318 | 5.635 ± 0.341 |
6.453 ± 0.581 | 5.726 ± 0.415 |
1.181 ± 0.145 | 4.938 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
