
Sewage-associated circular DNA virus-12
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
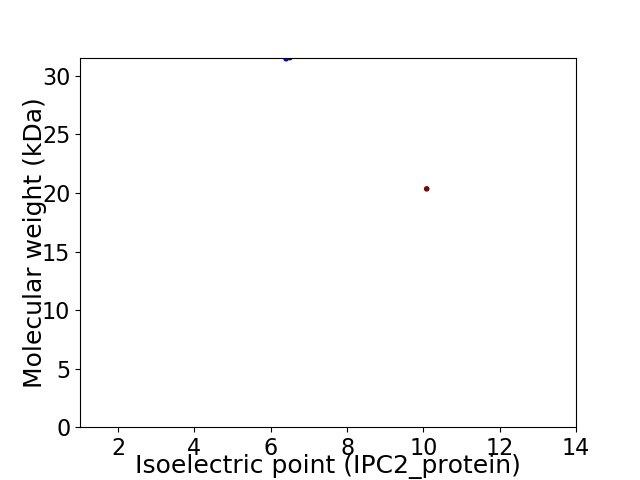
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J453|A0A075J453_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-12 OX=1519388 PE=4 SV=1
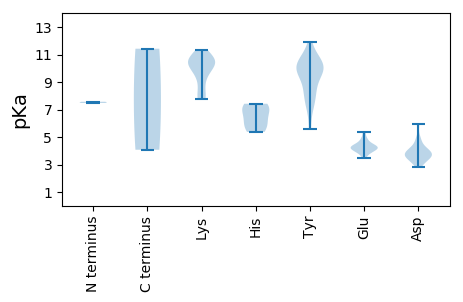
MM1 pKa = 7.48ALKK4 pKa = 10.4ARR6 pKa = 11.84YY7 pKa = 8.15WCFTLNNWCDD17 pKa = 3.61GEE19 pKa = 4.26LLEE22 pKa = 4.82IAKK25 pKa = 10.56FSGRR29 pKa = 11.84YY30 pKa = 5.61LTYY33 pKa = 9.95GFEE36 pKa = 4.33IGEE39 pKa = 4.44SGTPHH44 pKa = 5.81LQGYY48 pKa = 8.58CEE50 pKa = 4.22YY51 pKa = 9.75EE52 pKa = 4.14NPVTMGRR59 pKa = 11.84VKK61 pKa = 10.71KK62 pKa = 10.46DD63 pKa = 2.84ISDD66 pKa = 3.64RR67 pKa = 11.84CHH69 pKa = 7.16VEE71 pKa = 3.87VRR73 pKa = 11.84KK74 pKa = 8.62GTQLQAVSYY83 pKa = 8.5CHH85 pKa = 7.42KK86 pKa = 10.6DD87 pKa = 3.93GNWLEE92 pKa = 4.22YY93 pKa = 10.83GEE95 pKa = 4.43KK96 pKa = 10.44AAGQGHH102 pKa = 6.96RR103 pKa = 11.84SDD105 pKa = 4.79LDD107 pKa = 3.58DD108 pKa = 3.53VARR111 pKa = 11.84DD112 pKa = 3.82VLDD115 pKa = 3.99KK116 pKa = 11.05KK117 pKa = 11.33SLMEE121 pKa = 4.24IANDD125 pKa = 4.56HH126 pKa = 5.37PTSWIRR132 pKa = 11.84YY133 pKa = 7.42SRR135 pKa = 11.84GIQSLANSQPPPRR148 pKa = 11.84WRR150 pKa = 11.84DD151 pKa = 3.48VLVLVYY157 pKa = 9.92WGVTGTGKK165 pKa = 7.99TRR167 pKa = 11.84RR168 pKa = 11.84AMLEE172 pKa = 3.49PSAYY176 pKa = 9.72KK177 pKa = 10.64LNQNTNGTLWFDD189 pKa = 4.03GYY191 pKa = 10.97DD192 pKa = 3.69GQSTLVIDD200 pKa = 5.94DD201 pKa = 4.29FYY203 pKa = 11.9GWIKK207 pKa = 10.77YY208 pKa = 10.42GEE210 pKa = 4.82LLTLLDD216 pKa = 4.97GYY218 pKa = 9.6PYY220 pKa = 10.28RR221 pKa = 11.84CQVKK225 pKa = 10.41GGMAWARR232 pKa = 11.84WDD234 pKa = 3.7TVIITSNKK242 pKa = 9.7CPDD245 pKa = 3.16DD246 pKa = 4.1WYY248 pKa = 10.54PLVPDD253 pKa = 4.16TAALKK258 pKa = 10.48RR259 pKa = 11.84RR260 pKa = 11.84ISAIRR265 pKa = 11.84YY266 pKa = 7.13FEE268 pKa = 5.37PILDD272 pKa = 3.77QKK274 pKa = 11.42
MM1 pKa = 7.48ALKK4 pKa = 10.4ARR6 pKa = 11.84YY7 pKa = 8.15WCFTLNNWCDD17 pKa = 3.61GEE19 pKa = 4.26LLEE22 pKa = 4.82IAKK25 pKa = 10.56FSGRR29 pKa = 11.84YY30 pKa = 5.61LTYY33 pKa = 9.95GFEE36 pKa = 4.33IGEE39 pKa = 4.44SGTPHH44 pKa = 5.81LQGYY48 pKa = 8.58CEE50 pKa = 4.22YY51 pKa = 9.75EE52 pKa = 4.14NPVTMGRR59 pKa = 11.84VKK61 pKa = 10.71KK62 pKa = 10.46DD63 pKa = 2.84ISDD66 pKa = 3.64RR67 pKa = 11.84CHH69 pKa = 7.16VEE71 pKa = 3.87VRR73 pKa = 11.84KK74 pKa = 8.62GTQLQAVSYY83 pKa = 8.5CHH85 pKa = 7.42KK86 pKa = 10.6DD87 pKa = 3.93GNWLEE92 pKa = 4.22YY93 pKa = 10.83GEE95 pKa = 4.43KK96 pKa = 10.44AAGQGHH102 pKa = 6.96RR103 pKa = 11.84SDD105 pKa = 4.79LDD107 pKa = 3.58DD108 pKa = 3.53VARR111 pKa = 11.84DD112 pKa = 3.82VLDD115 pKa = 3.99KK116 pKa = 11.05KK117 pKa = 11.33SLMEE121 pKa = 4.24IANDD125 pKa = 4.56HH126 pKa = 5.37PTSWIRR132 pKa = 11.84YY133 pKa = 7.42SRR135 pKa = 11.84GIQSLANSQPPPRR148 pKa = 11.84WRR150 pKa = 11.84DD151 pKa = 3.48VLVLVYY157 pKa = 9.92WGVTGTGKK165 pKa = 7.99TRR167 pKa = 11.84RR168 pKa = 11.84AMLEE172 pKa = 3.49PSAYY176 pKa = 9.72KK177 pKa = 10.64LNQNTNGTLWFDD189 pKa = 4.03GYY191 pKa = 10.97DD192 pKa = 3.69GQSTLVIDD200 pKa = 5.94DD201 pKa = 4.29FYY203 pKa = 11.9GWIKK207 pKa = 10.77YY208 pKa = 10.42GEE210 pKa = 4.82LLTLLDD216 pKa = 4.97GYY218 pKa = 9.6PYY220 pKa = 10.28RR221 pKa = 11.84CQVKK225 pKa = 10.41GGMAWARR232 pKa = 11.84WDD234 pKa = 3.7TVIITSNKK242 pKa = 9.7CPDD245 pKa = 3.16DD246 pKa = 4.1WYY248 pKa = 10.54PLVPDD253 pKa = 4.16TAALKK258 pKa = 10.48RR259 pKa = 11.84RR260 pKa = 11.84ISAIRR265 pKa = 11.84YY266 pKa = 7.13FEE268 pKa = 5.37PILDD272 pKa = 3.77QKK274 pKa = 11.42
Molecular weight: 31.46 kDa
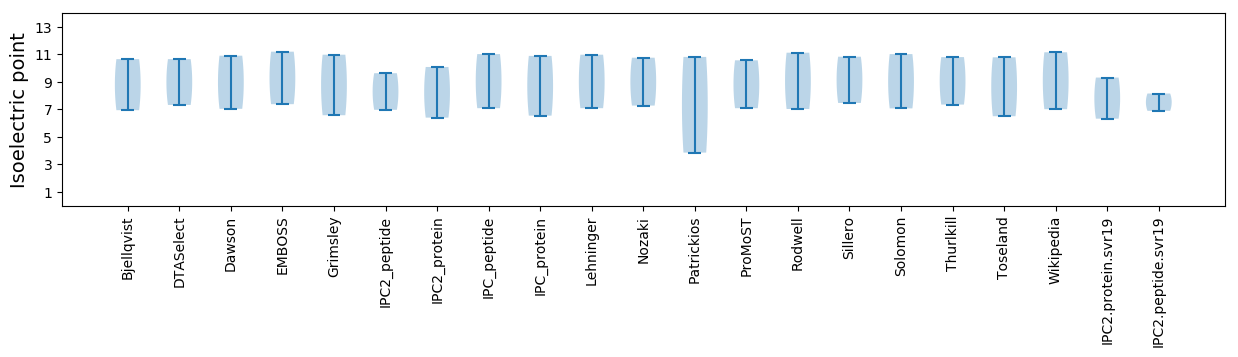
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J453|A0A075J453_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-12 OX=1519388 PE=4 SV=1
MM1 pKa = 7.57FLRR4 pKa = 11.84PQMPRR9 pKa = 11.84GYY11 pKa = 10.61YY12 pKa = 9.68KK13 pKa = 8.91PTYY16 pKa = 9.84RR17 pKa = 11.84KK18 pKa = 7.74RR19 pKa = 11.84TYY21 pKa = 10.52RR22 pKa = 11.84KK23 pKa = 7.73RR24 pKa = 11.84TYY26 pKa = 7.41TRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NTYY33 pKa = 9.58ARR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 7.86WSPSIKK43 pKa = 10.25YY44 pKa = 10.05KK45 pKa = 10.98SEE47 pKa = 3.92TKK49 pKa = 9.78WYY51 pKa = 10.06DD52 pKa = 3.11GTAGGGGIGNMPAAWNRR69 pKa = 11.84YY70 pKa = 8.71DD71 pKa = 4.07LSCPITQGVTSVSRR85 pKa = 11.84VGNNIIAKK93 pKa = 9.82SLGISFDD100 pKa = 3.36VTRR103 pKa = 11.84NASSSTPQQRR113 pKa = 11.84MRR115 pKa = 11.84FMVIDD120 pKa = 3.72YY121 pKa = 8.08PRR123 pKa = 11.84SEE125 pKa = 4.18GSIPAAAEE133 pKa = 4.26FFQTTGVFLPQRR145 pKa = 11.84NLSWVKK151 pKa = 9.94EE152 pKa = 3.95FHH154 pKa = 6.23VLADD158 pKa = 3.62RR159 pKa = 11.84VITVDD164 pKa = 3.18SASRR168 pKa = 11.84NSQMGKK174 pKa = 10.24AFPII178 pKa = 4.07
MM1 pKa = 7.57FLRR4 pKa = 11.84PQMPRR9 pKa = 11.84GYY11 pKa = 10.61YY12 pKa = 9.68KK13 pKa = 8.91PTYY16 pKa = 9.84RR17 pKa = 11.84KK18 pKa = 7.74RR19 pKa = 11.84TYY21 pKa = 10.52RR22 pKa = 11.84KK23 pKa = 7.73RR24 pKa = 11.84TYY26 pKa = 7.41TRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NTYY33 pKa = 9.58ARR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 7.86WSPSIKK43 pKa = 10.25YY44 pKa = 10.05KK45 pKa = 10.98SEE47 pKa = 3.92TKK49 pKa = 9.78WYY51 pKa = 10.06DD52 pKa = 3.11GTAGGGGIGNMPAAWNRR69 pKa = 11.84YY70 pKa = 8.71DD71 pKa = 4.07LSCPITQGVTSVSRR85 pKa = 11.84VGNNIIAKK93 pKa = 9.82SLGISFDD100 pKa = 3.36VTRR103 pKa = 11.84NASSSTPQQRR113 pKa = 11.84MRR115 pKa = 11.84FMVIDD120 pKa = 3.72YY121 pKa = 8.08PRR123 pKa = 11.84SEE125 pKa = 4.18GSIPAAAEE133 pKa = 4.26FFQTTGVFLPQRR145 pKa = 11.84NLSWVKK151 pKa = 9.94EE152 pKa = 3.95FHH154 pKa = 6.23VLADD158 pKa = 3.62RR159 pKa = 11.84VITVDD164 pKa = 3.18SASRR168 pKa = 11.84NSQMGKK174 pKa = 10.24AFPII178 pKa = 4.07
Molecular weight: 20.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
452 |
178 |
274 |
226.0 |
25.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.195 ± 0.326 | 1.77 ± 0.72 |
5.973 ± 1.552 | 3.761 ± 0.902 |
3.097 ± 0.833 | 8.186 ± 0.526 |
1.327 ± 0.456 | 5.088 ± 0.316 |
5.973 ± 0.123 | 6.858 ± 2.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.434 ± 0.559 | 3.982 ± 0.305 |
5.088 ± 0.651 | 3.761 ± 0.102 |
8.186 ± 1.483 | 6.858 ± 1.605 |
6.637 ± 0.732 | 5.531 ± 0.052 |
3.319 ± 0.639 | 5.973 ± 0.212 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
