
Beihai tombus-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
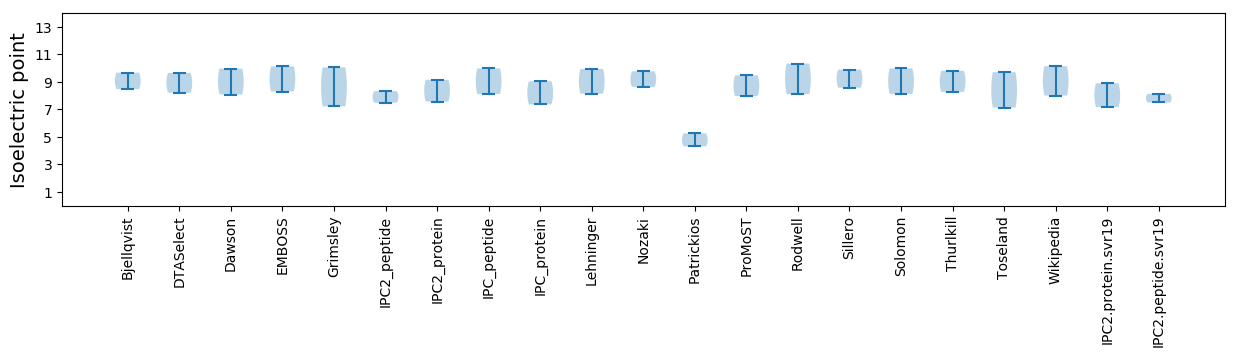
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
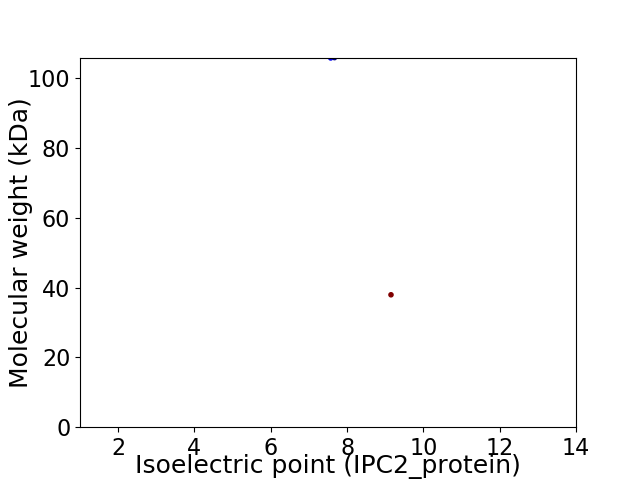
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFF2|A0A1L3KFF2_9VIRU RNA-directed RNA polymerase OS=Beihai tombus-like virus 15 OX=1922718 PE=4 SV=1
MM1 pKa = 7.76ALNLKK6 pKa = 8.8TNSFVLDD13 pKa = 3.93TEE15 pKa = 4.23QEE17 pKa = 3.9RR18 pKa = 11.84ALARR22 pKa = 11.84WGFNLKK28 pKa = 10.37SGTIPHH34 pKa = 6.23THH36 pKa = 6.69PVSALIRR43 pKa = 11.84NGFEE47 pKa = 3.8DD48 pKa = 4.99AVINQHH54 pKa = 5.15KK55 pKa = 10.08DD56 pKa = 3.01KK57 pKa = 10.62RR58 pKa = 11.84IKK60 pKa = 10.73DD61 pKa = 3.41VGGNYY66 pKa = 9.26ARR68 pKa = 11.84HH69 pKa = 6.14AARR72 pKa = 11.84HH73 pKa = 5.05QGFDD77 pKa = 3.0IHH79 pKa = 6.8SCCPLLSPDD88 pKa = 3.64DD89 pKa = 4.69LIRR92 pKa = 11.84EE93 pKa = 4.44VTRR96 pKa = 11.84DD97 pKa = 3.43MAVDD101 pKa = 3.91PDD103 pKa = 3.92CCRR106 pKa = 11.84KK107 pKa = 9.92RR108 pKa = 11.84FQDD111 pKa = 3.48CSRR114 pKa = 11.84PADD117 pKa = 3.15VYY119 pKa = 10.37MFNHH123 pKa = 5.29SMYY126 pKa = 10.3YY127 pKa = 9.82VEE129 pKa = 5.84PEE131 pKa = 4.2DD132 pKa = 4.05LACIPVGACIYY143 pKa = 10.05SLHH146 pKa = 6.83HH147 pKa = 7.19LYY149 pKa = 10.29QGPGTYY155 pKa = 9.93CAGEE159 pKa = 4.07MEE161 pKa = 4.56VTQDD165 pKa = 3.22EE166 pKa = 4.56QGVYY170 pKa = 9.43TVDD173 pKa = 2.82TRR175 pKa = 11.84GNNRR179 pKa = 11.84PYY181 pKa = 10.71KK182 pKa = 10.38HH183 pKa = 5.79KK184 pKa = 10.63QCWLTGSCVIPGKK197 pKa = 10.35DD198 pKa = 3.09GAVIAFTVLRR208 pKa = 11.84NYY210 pKa = 10.32EE211 pKa = 4.04SAHH214 pKa = 4.6VFKK217 pKa = 10.95GYY219 pKa = 9.78VYY221 pKa = 10.44KK222 pKa = 11.06VEE224 pKa = 4.08DD225 pKa = 3.86SYY227 pKa = 12.12RR228 pKa = 11.84LLSQPVFKK236 pKa = 10.31PYY238 pKa = 10.4DD239 pKa = 3.47IYY241 pKa = 10.83PSEE244 pKa = 5.34LEE246 pKa = 5.16DD247 pKa = 3.41ILINEE252 pKa = 4.72GTSQTIDD259 pKa = 2.83NRR261 pKa = 11.84TLHH264 pKa = 6.09NMASRR269 pKa = 11.84ARR271 pKa = 11.84TYY273 pKa = 11.59ALTTHH278 pKa = 6.94RR279 pKa = 11.84KK280 pKa = 7.5MPANIADD287 pKa = 4.22IYY289 pKa = 11.5ASAANKK295 pKa = 9.66AAEE298 pKa = 4.39INIKK302 pKa = 7.82TQSQLNKK309 pKa = 10.44EE310 pKa = 4.33KK311 pKa = 10.64IEE313 pKa = 4.01TSNRR317 pKa = 11.84LIAGLEE323 pKa = 3.85IDD325 pKa = 4.03TSQPHH330 pKa = 5.03IPIYY334 pKa = 10.36RR335 pKa = 11.84RR336 pKa = 11.84VGGILWTLASGVVRR350 pKa = 11.84TTAYY354 pKa = 9.67TSLDD358 pKa = 3.61YY359 pKa = 10.69MKK361 pKa = 10.85AGALLPAKK369 pKa = 10.2AIIYY373 pKa = 5.08TTSRR377 pKa = 11.84IRR379 pKa = 11.84QCLTYY384 pKa = 10.43EE385 pKa = 4.03NNDD388 pKa = 3.51YY389 pKa = 11.02VVPTCTFVPLNSWEE403 pKa = 4.23NEE405 pKa = 3.83LTSILLRR412 pKa = 11.84AVPDD416 pKa = 3.96TVTTTIDD423 pKa = 3.4NKK425 pKa = 10.51IMEE428 pKa = 4.86SADD431 pKa = 3.61KK432 pKa = 10.33LAKK435 pKa = 10.55LIGYY439 pKa = 9.14VDD441 pKa = 3.76APMHH445 pKa = 5.78FDD447 pKa = 3.03EE448 pKa = 4.67WVEE451 pKa = 3.97RR452 pKa = 11.84YY453 pKa = 9.17PPARR457 pKa = 11.84QEE459 pKa = 4.3QLRR462 pKa = 11.84KK463 pKa = 9.78AHH465 pKa = 5.88YY466 pKa = 9.95KK467 pKa = 9.57PLNHH471 pKa = 6.3NVDD474 pKa = 3.39MFIKK478 pKa = 10.07IEE480 pKa = 3.85QLDD483 pKa = 4.06DD484 pKa = 3.65QKK486 pKa = 11.28HH487 pKa = 5.08PRR489 pKa = 11.84AIQARR494 pKa = 11.84HH495 pKa = 6.39DD496 pKa = 4.02SYY498 pKa = 11.41KK499 pKa = 10.9SKK501 pKa = 10.66LGPWVARR508 pKa = 11.84LEE510 pKa = 4.24KK511 pKa = 10.49RR512 pKa = 11.84CIEE515 pKa = 4.13ALPNFVKK522 pKa = 10.7KK523 pKa = 11.02LNDD526 pKa = 3.16EE527 pKa = 3.96QRR529 pKa = 11.84AEE531 pKa = 4.06KK532 pKa = 10.51VAEE535 pKa = 4.44LKK537 pKa = 10.87CKK539 pKa = 10.11ADD541 pKa = 3.99NIIEE545 pKa = 4.03IDD547 pKa = 3.46FTRR550 pKa = 11.84FDD552 pKa = 4.13RR553 pKa = 11.84NCTKK557 pKa = 10.44EE558 pKa = 4.06LLSATEE564 pKa = 4.04HH565 pKa = 6.07YY566 pKa = 10.37IYY568 pKa = 11.02DD569 pKa = 3.51KK570 pKa = 11.29VFPKK574 pKa = 10.53EE575 pKa = 3.86IASLLHH581 pKa = 5.5LQLRR585 pKa = 11.84NKK587 pKa = 9.08VRR589 pKa = 11.84SARR592 pKa = 11.84GYY594 pKa = 7.79TYY596 pKa = 10.54SVEE599 pKa = 4.15GTRR602 pKa = 11.84MSGDD606 pKa = 3.37VNTSIGNCLIVLCLSLAAGLEE627 pKa = 4.15LEE629 pKa = 4.73NILVEE634 pKa = 4.47GDD636 pKa = 3.78DD637 pKa = 4.46MIAAASDD644 pKa = 2.96ITLQKK649 pKa = 10.85FSDD652 pKa = 4.04EE653 pKa = 4.43VIKK656 pKa = 10.02ATGMIPKK663 pKa = 8.89TVVTHH668 pKa = 6.92RR669 pKa = 11.84GGSFCSRR676 pKa = 11.84YY677 pKa = 10.37DD678 pKa = 3.71VIDD681 pKa = 3.75SEE683 pKa = 4.67GKK685 pKa = 8.32PRR687 pKa = 11.84RR688 pKa = 11.84VRR690 pKa = 11.84HH691 pKa = 5.5PLRR694 pKa = 11.84DD695 pKa = 3.24ITRR698 pKa = 11.84YY699 pKa = 10.42GYY701 pKa = 8.0TLHH704 pKa = 7.73GEE706 pKa = 4.23DD707 pKa = 3.8RR708 pKa = 11.84LEE710 pKa = 3.98RR711 pKa = 11.84AQRR714 pKa = 11.84HH715 pKa = 4.14YY716 pKa = 10.74KK717 pKa = 9.41EE718 pKa = 4.36WVGVPMLGPVYY729 pKa = 10.59TNILKK734 pKa = 9.91QLQPDD739 pKa = 3.74VQIPEE744 pKa = 3.93IEE746 pKa = 3.97ITAEE750 pKa = 3.75ARR752 pKa = 11.84VSFARR757 pKa = 11.84TFDD760 pKa = 3.42ISIKK764 pKa = 7.47TQQLFEE770 pKa = 4.31EE771 pKa = 5.16DD772 pKa = 2.93KK773 pKa = 10.43SQRR776 pKa = 11.84ARR778 pKa = 11.84IYY780 pKa = 11.33NDD782 pKa = 2.96LTQEE786 pKa = 4.42KK787 pKa = 9.41IYY789 pKa = 9.48KK790 pKa = 8.36TALTEE795 pKa = 4.06AVEE798 pKa = 4.39TPIIGPGDD806 pKa = 3.91SVSQISGTPGSSEE819 pKa = 3.97TGHH822 pKa = 7.4DD823 pKa = 3.64KK824 pKa = 11.23LQVPTWNIRR833 pKa = 11.84TGSLGSTSKK842 pKa = 10.67NVRR845 pKa = 11.84DD846 pKa = 4.02VQVKK850 pKa = 9.91GPSEE854 pKa = 4.24SGLQNSLRR862 pKa = 11.84DD863 pKa = 3.69DD864 pKa = 3.74NKK866 pKa = 10.64RR867 pKa = 11.84GNTHH871 pKa = 6.42RR872 pKa = 11.84NRR874 pKa = 11.84LRR876 pKa = 11.84RR877 pKa = 11.84HH878 pKa = 5.4RR879 pKa = 11.84RR880 pKa = 11.84QARR883 pKa = 11.84LHH885 pKa = 5.1GHH887 pKa = 6.0GSIVSKK893 pKa = 11.39DD894 pKa = 3.58NVTGLAGLPGGSSKK908 pKa = 10.81RR909 pKa = 11.84SGNEE913 pKa = 3.06TEE915 pKa = 4.58MAMHH919 pKa = 7.47CDD921 pKa = 3.32PSKK924 pKa = 10.18PNSSIGQYY932 pKa = 9.59SIRR935 pKa = 11.84NPSTT939 pKa = 3.01
MM1 pKa = 7.76ALNLKK6 pKa = 8.8TNSFVLDD13 pKa = 3.93TEE15 pKa = 4.23QEE17 pKa = 3.9RR18 pKa = 11.84ALARR22 pKa = 11.84WGFNLKK28 pKa = 10.37SGTIPHH34 pKa = 6.23THH36 pKa = 6.69PVSALIRR43 pKa = 11.84NGFEE47 pKa = 3.8DD48 pKa = 4.99AVINQHH54 pKa = 5.15KK55 pKa = 10.08DD56 pKa = 3.01KK57 pKa = 10.62RR58 pKa = 11.84IKK60 pKa = 10.73DD61 pKa = 3.41VGGNYY66 pKa = 9.26ARR68 pKa = 11.84HH69 pKa = 6.14AARR72 pKa = 11.84HH73 pKa = 5.05QGFDD77 pKa = 3.0IHH79 pKa = 6.8SCCPLLSPDD88 pKa = 3.64DD89 pKa = 4.69LIRR92 pKa = 11.84EE93 pKa = 4.44VTRR96 pKa = 11.84DD97 pKa = 3.43MAVDD101 pKa = 3.91PDD103 pKa = 3.92CCRR106 pKa = 11.84KK107 pKa = 9.92RR108 pKa = 11.84FQDD111 pKa = 3.48CSRR114 pKa = 11.84PADD117 pKa = 3.15VYY119 pKa = 10.37MFNHH123 pKa = 5.29SMYY126 pKa = 10.3YY127 pKa = 9.82VEE129 pKa = 5.84PEE131 pKa = 4.2DD132 pKa = 4.05LACIPVGACIYY143 pKa = 10.05SLHH146 pKa = 6.83HH147 pKa = 7.19LYY149 pKa = 10.29QGPGTYY155 pKa = 9.93CAGEE159 pKa = 4.07MEE161 pKa = 4.56VTQDD165 pKa = 3.22EE166 pKa = 4.56QGVYY170 pKa = 9.43TVDD173 pKa = 2.82TRR175 pKa = 11.84GNNRR179 pKa = 11.84PYY181 pKa = 10.71KK182 pKa = 10.38HH183 pKa = 5.79KK184 pKa = 10.63QCWLTGSCVIPGKK197 pKa = 10.35DD198 pKa = 3.09GAVIAFTVLRR208 pKa = 11.84NYY210 pKa = 10.32EE211 pKa = 4.04SAHH214 pKa = 4.6VFKK217 pKa = 10.95GYY219 pKa = 9.78VYY221 pKa = 10.44KK222 pKa = 11.06VEE224 pKa = 4.08DD225 pKa = 3.86SYY227 pKa = 12.12RR228 pKa = 11.84LLSQPVFKK236 pKa = 10.31PYY238 pKa = 10.4DD239 pKa = 3.47IYY241 pKa = 10.83PSEE244 pKa = 5.34LEE246 pKa = 5.16DD247 pKa = 3.41ILINEE252 pKa = 4.72GTSQTIDD259 pKa = 2.83NRR261 pKa = 11.84TLHH264 pKa = 6.09NMASRR269 pKa = 11.84ARR271 pKa = 11.84TYY273 pKa = 11.59ALTTHH278 pKa = 6.94RR279 pKa = 11.84KK280 pKa = 7.5MPANIADD287 pKa = 4.22IYY289 pKa = 11.5ASAANKK295 pKa = 9.66AAEE298 pKa = 4.39INIKK302 pKa = 7.82TQSQLNKK309 pKa = 10.44EE310 pKa = 4.33KK311 pKa = 10.64IEE313 pKa = 4.01TSNRR317 pKa = 11.84LIAGLEE323 pKa = 3.85IDD325 pKa = 4.03TSQPHH330 pKa = 5.03IPIYY334 pKa = 10.36RR335 pKa = 11.84RR336 pKa = 11.84VGGILWTLASGVVRR350 pKa = 11.84TTAYY354 pKa = 9.67TSLDD358 pKa = 3.61YY359 pKa = 10.69MKK361 pKa = 10.85AGALLPAKK369 pKa = 10.2AIIYY373 pKa = 5.08TTSRR377 pKa = 11.84IRR379 pKa = 11.84QCLTYY384 pKa = 10.43EE385 pKa = 4.03NNDD388 pKa = 3.51YY389 pKa = 11.02VVPTCTFVPLNSWEE403 pKa = 4.23NEE405 pKa = 3.83LTSILLRR412 pKa = 11.84AVPDD416 pKa = 3.96TVTTTIDD423 pKa = 3.4NKK425 pKa = 10.51IMEE428 pKa = 4.86SADD431 pKa = 3.61KK432 pKa = 10.33LAKK435 pKa = 10.55LIGYY439 pKa = 9.14VDD441 pKa = 3.76APMHH445 pKa = 5.78FDD447 pKa = 3.03EE448 pKa = 4.67WVEE451 pKa = 3.97RR452 pKa = 11.84YY453 pKa = 9.17PPARR457 pKa = 11.84QEE459 pKa = 4.3QLRR462 pKa = 11.84KK463 pKa = 9.78AHH465 pKa = 5.88YY466 pKa = 9.95KK467 pKa = 9.57PLNHH471 pKa = 6.3NVDD474 pKa = 3.39MFIKK478 pKa = 10.07IEE480 pKa = 3.85QLDD483 pKa = 4.06DD484 pKa = 3.65QKK486 pKa = 11.28HH487 pKa = 5.08PRR489 pKa = 11.84AIQARR494 pKa = 11.84HH495 pKa = 6.39DD496 pKa = 4.02SYY498 pKa = 11.41KK499 pKa = 10.9SKK501 pKa = 10.66LGPWVARR508 pKa = 11.84LEE510 pKa = 4.24KK511 pKa = 10.49RR512 pKa = 11.84CIEE515 pKa = 4.13ALPNFVKK522 pKa = 10.7KK523 pKa = 11.02LNDD526 pKa = 3.16EE527 pKa = 3.96QRR529 pKa = 11.84AEE531 pKa = 4.06KK532 pKa = 10.51VAEE535 pKa = 4.44LKK537 pKa = 10.87CKK539 pKa = 10.11ADD541 pKa = 3.99NIIEE545 pKa = 4.03IDD547 pKa = 3.46FTRR550 pKa = 11.84FDD552 pKa = 4.13RR553 pKa = 11.84NCTKK557 pKa = 10.44EE558 pKa = 4.06LLSATEE564 pKa = 4.04HH565 pKa = 6.07YY566 pKa = 10.37IYY568 pKa = 11.02DD569 pKa = 3.51KK570 pKa = 11.29VFPKK574 pKa = 10.53EE575 pKa = 3.86IASLLHH581 pKa = 5.5LQLRR585 pKa = 11.84NKK587 pKa = 9.08VRR589 pKa = 11.84SARR592 pKa = 11.84GYY594 pKa = 7.79TYY596 pKa = 10.54SVEE599 pKa = 4.15GTRR602 pKa = 11.84MSGDD606 pKa = 3.37VNTSIGNCLIVLCLSLAAGLEE627 pKa = 4.15LEE629 pKa = 4.73NILVEE634 pKa = 4.47GDD636 pKa = 3.78DD637 pKa = 4.46MIAAASDD644 pKa = 2.96ITLQKK649 pKa = 10.85FSDD652 pKa = 4.04EE653 pKa = 4.43VIKK656 pKa = 10.02ATGMIPKK663 pKa = 8.89TVVTHH668 pKa = 6.92RR669 pKa = 11.84GGSFCSRR676 pKa = 11.84YY677 pKa = 10.37DD678 pKa = 3.71VIDD681 pKa = 3.75SEE683 pKa = 4.67GKK685 pKa = 8.32PRR687 pKa = 11.84RR688 pKa = 11.84VRR690 pKa = 11.84HH691 pKa = 5.5PLRR694 pKa = 11.84DD695 pKa = 3.24ITRR698 pKa = 11.84YY699 pKa = 10.42GYY701 pKa = 8.0TLHH704 pKa = 7.73GEE706 pKa = 4.23DD707 pKa = 3.8RR708 pKa = 11.84LEE710 pKa = 3.98RR711 pKa = 11.84AQRR714 pKa = 11.84HH715 pKa = 4.14YY716 pKa = 10.74KK717 pKa = 9.41EE718 pKa = 4.36WVGVPMLGPVYY729 pKa = 10.59TNILKK734 pKa = 9.91QLQPDD739 pKa = 3.74VQIPEE744 pKa = 3.93IEE746 pKa = 3.97ITAEE750 pKa = 3.75ARR752 pKa = 11.84VSFARR757 pKa = 11.84TFDD760 pKa = 3.42ISIKK764 pKa = 7.47TQQLFEE770 pKa = 4.31EE771 pKa = 5.16DD772 pKa = 2.93KK773 pKa = 10.43SQRR776 pKa = 11.84ARR778 pKa = 11.84IYY780 pKa = 11.33NDD782 pKa = 2.96LTQEE786 pKa = 4.42KK787 pKa = 9.41IYY789 pKa = 9.48KK790 pKa = 8.36TALTEE795 pKa = 4.06AVEE798 pKa = 4.39TPIIGPGDD806 pKa = 3.91SVSQISGTPGSSEE819 pKa = 3.97TGHH822 pKa = 7.4DD823 pKa = 3.64KK824 pKa = 11.23LQVPTWNIRR833 pKa = 11.84TGSLGSTSKK842 pKa = 10.67NVRR845 pKa = 11.84DD846 pKa = 4.02VQVKK850 pKa = 9.91GPSEE854 pKa = 4.24SGLQNSLRR862 pKa = 11.84DD863 pKa = 3.69DD864 pKa = 3.74NKK866 pKa = 10.64RR867 pKa = 11.84GNTHH871 pKa = 6.42RR872 pKa = 11.84NRR874 pKa = 11.84LRR876 pKa = 11.84RR877 pKa = 11.84HH878 pKa = 5.4RR879 pKa = 11.84RR880 pKa = 11.84QARR883 pKa = 11.84LHH885 pKa = 5.1GHH887 pKa = 6.0GSIVSKK893 pKa = 11.39DD894 pKa = 3.58NVTGLAGLPGGSSKK908 pKa = 10.81RR909 pKa = 11.84SGNEE913 pKa = 3.06TEE915 pKa = 4.58MAMHH919 pKa = 7.47CDD921 pKa = 3.32PSKK924 pKa = 10.18PNSSIGQYY932 pKa = 9.59SIRR935 pKa = 11.84NPSTT939 pKa = 3.01
Molecular weight: 105.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFF2|A0A1L3KFF2_9VIRU RNA-directed RNA polymerase OS=Beihai tombus-like virus 15 OX=1922718 PE=4 SV=1
MM1 pKa = 7.34YY2 pKa = 10.0KK3 pKa = 9.8YY4 pKa = 10.2QKK6 pKa = 10.69SKK8 pKa = 10.95LPQKK12 pKa = 10.23LAYY15 pKa = 9.11PSQEE19 pKa = 3.8HH20 pKa = 5.76STSQSKK26 pKa = 10.55HH27 pKa = 5.6SNSSKK32 pKa = 10.38KK33 pKa = 10.12ISPKK37 pKa = 9.33EE38 pKa = 3.87HH39 pKa = 6.34VSTMTSPKK47 pKa = 9.92RR48 pKa = 11.84KK49 pKa = 7.26STKK52 pKa = 9.54RR53 pKa = 11.84PSPKK57 pKa = 9.08RR58 pKa = 11.84SKK60 pKa = 10.62RR61 pKa = 11.84PSLAPGTLLVKK72 pKa = 10.48YY73 pKa = 9.91RR74 pKa = 11.84EE75 pKa = 3.96LLAPVKK81 pKa = 10.55QGTTSYY87 pKa = 10.76KK88 pKa = 10.01FRR90 pKa = 11.84PGTSGLAHH98 pKa = 7.2LDD100 pKa = 3.13QRR102 pKa = 11.84ARR104 pKa = 11.84MYY106 pKa = 11.04EE107 pKa = 4.06MYY109 pKa = 10.5RR110 pKa = 11.84LRR112 pKa = 11.84GPVRR116 pKa = 11.84VGYY119 pKa = 8.25KK120 pKa = 8.57TASGTTTNGEE130 pKa = 3.88ILIGIDD136 pKa = 4.86FDD138 pKa = 3.97ATDD141 pKa = 4.41DD142 pKa = 3.79RR143 pKa = 11.84LDD145 pKa = 3.8YY146 pKa = 10.54MGTAALSPKK155 pKa = 9.52TMSPVWRR162 pKa = 11.84DD163 pKa = 3.47CQVVVPKK170 pKa = 10.63DD171 pKa = 3.08RR172 pKa = 11.84AMKK175 pKa = 10.09QKK177 pKa = 9.25WLCTATRR184 pKa = 11.84ASQTVASDD192 pKa = 3.24NTAFAIQVHH201 pKa = 5.69NSGPTGCGSLWVEE214 pKa = 4.3YY215 pKa = 9.77YY216 pKa = 11.08LEE218 pKa = 4.4FMSPCVNSEE227 pKa = 4.23DD228 pKa = 4.05VGIAYY233 pKa = 7.56ITTSGNGGGHH243 pKa = 6.46GGSVVGNPGLVPGGHH258 pKa = 6.11SKK260 pKa = 11.04AFAIPDD266 pKa = 3.46ALMPAFNEE274 pKa = 4.07ACKK277 pKa = 10.52AFPDD281 pKa = 3.65VMKK284 pKa = 10.31TVLVPDD290 pKa = 4.85GAPGGHH296 pKa = 6.24SYY298 pKa = 11.22INLEE302 pKa = 3.85NKK304 pKa = 9.83GYY306 pKa = 10.45KK307 pKa = 9.41DD308 pKa = 3.93AYY310 pKa = 9.89VSLPSTHH317 pKa = 6.56WPSSQQISFWFAAPSVAILVRR338 pKa = 11.84YY339 pKa = 8.95LLNKK343 pKa = 9.25NHH345 pKa = 6.63SSVV348 pKa = 3.16
MM1 pKa = 7.34YY2 pKa = 10.0KK3 pKa = 9.8YY4 pKa = 10.2QKK6 pKa = 10.69SKK8 pKa = 10.95LPQKK12 pKa = 10.23LAYY15 pKa = 9.11PSQEE19 pKa = 3.8HH20 pKa = 5.76STSQSKK26 pKa = 10.55HH27 pKa = 5.6SNSSKK32 pKa = 10.38KK33 pKa = 10.12ISPKK37 pKa = 9.33EE38 pKa = 3.87HH39 pKa = 6.34VSTMTSPKK47 pKa = 9.92RR48 pKa = 11.84KK49 pKa = 7.26STKK52 pKa = 9.54RR53 pKa = 11.84PSPKK57 pKa = 9.08RR58 pKa = 11.84SKK60 pKa = 10.62RR61 pKa = 11.84PSLAPGTLLVKK72 pKa = 10.48YY73 pKa = 9.91RR74 pKa = 11.84EE75 pKa = 3.96LLAPVKK81 pKa = 10.55QGTTSYY87 pKa = 10.76KK88 pKa = 10.01FRR90 pKa = 11.84PGTSGLAHH98 pKa = 7.2LDD100 pKa = 3.13QRR102 pKa = 11.84ARR104 pKa = 11.84MYY106 pKa = 11.04EE107 pKa = 4.06MYY109 pKa = 10.5RR110 pKa = 11.84LRR112 pKa = 11.84GPVRR116 pKa = 11.84VGYY119 pKa = 8.25KK120 pKa = 8.57TASGTTTNGEE130 pKa = 3.88ILIGIDD136 pKa = 4.86FDD138 pKa = 3.97ATDD141 pKa = 4.41DD142 pKa = 3.79RR143 pKa = 11.84LDD145 pKa = 3.8YY146 pKa = 10.54MGTAALSPKK155 pKa = 9.52TMSPVWRR162 pKa = 11.84DD163 pKa = 3.47CQVVVPKK170 pKa = 10.63DD171 pKa = 3.08RR172 pKa = 11.84AMKK175 pKa = 10.09QKK177 pKa = 9.25WLCTATRR184 pKa = 11.84ASQTVASDD192 pKa = 3.24NTAFAIQVHH201 pKa = 5.69NSGPTGCGSLWVEE214 pKa = 4.3YY215 pKa = 9.77YY216 pKa = 11.08LEE218 pKa = 4.4FMSPCVNSEE227 pKa = 4.23DD228 pKa = 4.05VGIAYY233 pKa = 7.56ITTSGNGGGHH243 pKa = 6.46GGSVVGNPGLVPGGHH258 pKa = 6.11SKK260 pKa = 11.04AFAIPDD266 pKa = 3.46ALMPAFNEE274 pKa = 4.07ACKK277 pKa = 10.52AFPDD281 pKa = 3.65VMKK284 pKa = 10.31TVLVPDD290 pKa = 4.85GAPGGHH296 pKa = 6.24SYY298 pKa = 11.22INLEE302 pKa = 3.85NKK304 pKa = 9.83GYY306 pKa = 10.45KK307 pKa = 9.41DD308 pKa = 3.93AYY310 pKa = 9.89VSLPSTHH317 pKa = 6.56WPSSQQISFWFAAPSVAILVRR338 pKa = 11.84YY339 pKa = 8.95LLNKK343 pKa = 9.25NHH345 pKa = 6.63SSVV348 pKa = 3.16
Molecular weight: 37.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1287 |
348 |
939 |
643.5 |
71.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.148 ± 0.291 | 1.865 ± 0.204 |
5.594 ± 0.749 | 5.128 ± 1.074 |
2.331 ± 0.122 | 6.527 ± 0.724 |
3.186 ± 0.149 | 5.828 ± 1.27 |
6.371 ± 0.661 | 7.615 ± 0.342 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.37 | 4.429 ± 0.467 |
5.517 ± 0.931 | 3.73 ± 0.134 |
6.449 ± 0.882 | 7.848 ± 1.463 |
6.76 ± 0.065 | 6.294 ± 0.287 |
1.01 ± 0.203 | 4.274 ± 0.154 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
