
Sewage-associated circular DNA virus-8
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
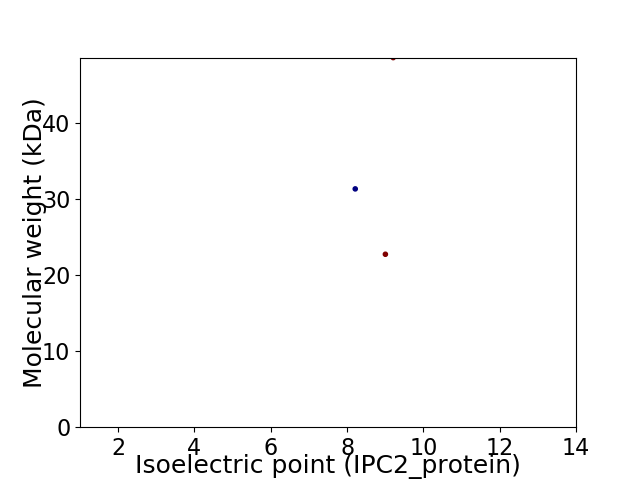
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J186|A0A075J186_9VIRU Uncharacterized protein OS=Sewage-associated circular DNA virus-8 OX=1519397 PE=4 SV=1
MM1 pKa = 7.19SQDD4 pKa = 4.79PLWKK8 pKa = 8.75MQGRR12 pKa = 11.84YY13 pKa = 7.51WMLTAPAASFTPYY26 pKa = 10.61LPPTVLYY33 pKa = 10.35IRR35 pKa = 11.84GQLEE39 pKa = 3.78TAASGFSHH47 pKa = 6.36WQLLVITGKK56 pKa = 9.97VRR58 pKa = 11.84GSKK61 pKa = 10.39LKK63 pKa = 9.97TIFGDD68 pKa = 3.83DD69 pKa = 3.31VHH71 pKa = 8.76IEE73 pKa = 4.12LTRR76 pKa = 11.84SAAADD81 pKa = 3.77DD82 pKa = 4.54YY83 pKa = 11.28VWKK86 pKa = 10.46DD87 pKa = 3.19DD88 pKa = 3.88TYY90 pKa = 11.14VDD92 pKa = 3.52GTRR95 pKa = 11.84FEE97 pKa = 4.4LGRR100 pKa = 11.84RR101 pKa = 11.84PKK103 pKa = 10.19QLSKK107 pKa = 8.32TTDD110 pKa = 2.67WDD112 pKa = 4.93DD113 pKa = 2.86IWKK116 pKa = 10.27SAVDD120 pKa = 4.06GNLEE124 pKa = 4.52DD125 pKa = 3.92IEE127 pKa = 4.66SHH129 pKa = 5.59VRR131 pKa = 11.84VRR133 pKa = 11.84YY134 pKa = 9.25YY135 pKa = 11.22NSLSRR140 pKa = 11.84IRR142 pKa = 11.84QDD144 pKa = 3.03HH145 pKa = 6.5LRR147 pKa = 11.84PIPLEE152 pKa = 4.17RR153 pKa = 11.84SCSVYY158 pKa = 9.73WGRR161 pKa = 11.84TGTGKK166 pKa = 9.99SRR168 pKa = 11.84RR169 pKa = 11.84AWEE172 pKa = 4.11EE173 pKa = 3.42ASMDD177 pKa = 4.16AYY179 pKa = 10.06PKK181 pKa = 10.53NPRR184 pKa = 11.84TKK186 pKa = 9.85FWDD189 pKa = 4.38GYY191 pKa = 9.67RR192 pKa = 11.84GHH194 pKa = 6.02EE195 pKa = 3.93HH196 pKa = 5.99VVIDD200 pKa = 4.17EE201 pKa = 4.11FRR203 pKa = 11.84GGIDD207 pKa = 3.14IAYY210 pKa = 7.76MLQWLDD216 pKa = 3.57RR217 pKa = 11.84YY218 pKa = 9.78PVIVEE223 pKa = 4.03VKK225 pKa = 10.35GSSTVLRR232 pKa = 11.84ATRR235 pKa = 11.84IWITSNIDD243 pKa = 3.03PRR245 pKa = 11.84DD246 pKa = 3.76WYY248 pKa = 10.27PLADD252 pKa = 4.06QEE254 pKa = 4.92TKK256 pKa = 10.77DD257 pKa = 3.45ALLRR261 pKa = 11.84RR262 pKa = 11.84LNITHH267 pKa = 7.31FLL269 pKa = 3.74
MM1 pKa = 7.19SQDD4 pKa = 4.79PLWKK8 pKa = 8.75MQGRR12 pKa = 11.84YY13 pKa = 7.51WMLTAPAASFTPYY26 pKa = 10.61LPPTVLYY33 pKa = 10.35IRR35 pKa = 11.84GQLEE39 pKa = 3.78TAASGFSHH47 pKa = 6.36WQLLVITGKK56 pKa = 9.97VRR58 pKa = 11.84GSKK61 pKa = 10.39LKK63 pKa = 9.97TIFGDD68 pKa = 3.83DD69 pKa = 3.31VHH71 pKa = 8.76IEE73 pKa = 4.12LTRR76 pKa = 11.84SAAADD81 pKa = 3.77DD82 pKa = 4.54YY83 pKa = 11.28VWKK86 pKa = 10.46DD87 pKa = 3.19DD88 pKa = 3.88TYY90 pKa = 11.14VDD92 pKa = 3.52GTRR95 pKa = 11.84FEE97 pKa = 4.4LGRR100 pKa = 11.84RR101 pKa = 11.84PKK103 pKa = 10.19QLSKK107 pKa = 8.32TTDD110 pKa = 2.67WDD112 pKa = 4.93DD113 pKa = 2.86IWKK116 pKa = 10.27SAVDD120 pKa = 4.06GNLEE124 pKa = 4.52DD125 pKa = 3.92IEE127 pKa = 4.66SHH129 pKa = 5.59VRR131 pKa = 11.84VRR133 pKa = 11.84YY134 pKa = 9.25YY135 pKa = 11.22NSLSRR140 pKa = 11.84IRR142 pKa = 11.84QDD144 pKa = 3.03HH145 pKa = 6.5LRR147 pKa = 11.84PIPLEE152 pKa = 4.17RR153 pKa = 11.84SCSVYY158 pKa = 9.73WGRR161 pKa = 11.84TGTGKK166 pKa = 9.99SRR168 pKa = 11.84RR169 pKa = 11.84AWEE172 pKa = 4.11EE173 pKa = 3.42ASMDD177 pKa = 4.16AYY179 pKa = 10.06PKK181 pKa = 10.53NPRR184 pKa = 11.84TKK186 pKa = 9.85FWDD189 pKa = 4.38GYY191 pKa = 9.67RR192 pKa = 11.84GHH194 pKa = 6.02EE195 pKa = 3.93HH196 pKa = 5.99VVIDD200 pKa = 4.17EE201 pKa = 4.11FRR203 pKa = 11.84GGIDD207 pKa = 3.14IAYY210 pKa = 7.76MLQWLDD216 pKa = 3.57RR217 pKa = 11.84YY218 pKa = 9.78PVIVEE223 pKa = 4.03VKK225 pKa = 10.35GSSTVLRR232 pKa = 11.84ATRR235 pKa = 11.84IWITSNIDD243 pKa = 3.03PRR245 pKa = 11.84DD246 pKa = 3.76WYY248 pKa = 10.27PLADD252 pKa = 4.06QEE254 pKa = 4.92TKK256 pKa = 10.77DD257 pKa = 3.45ALLRR261 pKa = 11.84RR262 pKa = 11.84LNITHH267 pKa = 7.31FLL269 pKa = 3.74
Molecular weight: 31.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J472|A0A075J472_9VIRU Uncharacterized protein OS=Sewage-associated circular DNA virus-8 OX=1519397 PE=4 SV=1
MM1 pKa = 7.51SCNSTLQVLFQPGILRR17 pKa = 11.84GSSPIFSIDD26 pKa = 3.21TLNTFDD32 pKa = 4.85PLLTEE37 pKa = 4.65LVPSFISANRR47 pKa = 11.84AARR50 pKa = 11.84SPLGSRR56 pKa = 11.84AVTFFGAKK64 pKa = 8.38TKK66 pKa = 10.76NFSLYY71 pKa = 10.87ARR73 pKa = 11.84LEE75 pKa = 4.04LFLNVTWQLEE85 pKa = 4.54SITTIFKK92 pKa = 9.79PSLWKK97 pKa = 9.19STPEE101 pKa = 4.05SYY103 pKa = 10.52PGTEE107 pKa = 4.27DD108 pKa = 3.61DD109 pKa = 3.96GSVRR113 pKa = 11.84FLHH116 pKa = 6.6KK117 pKa = 10.36IEE119 pKa = 4.3AASLNEE125 pKa = 3.84FGVNAKK131 pKa = 7.62GTPKK135 pKa = 10.67YY136 pKa = 10.18STFCLLSNTLPPNSGVPPNHH156 pKa = 6.74RR157 pKa = 11.84LEE159 pKa = 4.42TTFFVYY165 pKa = 10.59LPVTSPGANNRR176 pKa = 11.84LIASFMAGFNLTTFFRR192 pKa = 11.84FCLRR196 pKa = 11.84TFPRR200 pKa = 11.84RR201 pKa = 11.84TLPAA205 pKa = 4.75
MM1 pKa = 7.51SCNSTLQVLFQPGILRR17 pKa = 11.84GSSPIFSIDD26 pKa = 3.21TLNTFDD32 pKa = 4.85PLLTEE37 pKa = 4.65LVPSFISANRR47 pKa = 11.84AARR50 pKa = 11.84SPLGSRR56 pKa = 11.84AVTFFGAKK64 pKa = 8.38TKK66 pKa = 10.76NFSLYY71 pKa = 10.87ARR73 pKa = 11.84LEE75 pKa = 4.04LFLNVTWQLEE85 pKa = 4.54SITTIFKK92 pKa = 9.79PSLWKK97 pKa = 9.19STPEE101 pKa = 4.05SYY103 pKa = 10.52PGTEE107 pKa = 4.27DD108 pKa = 3.61DD109 pKa = 3.96GSVRR113 pKa = 11.84FLHH116 pKa = 6.6KK117 pKa = 10.36IEE119 pKa = 4.3AASLNEE125 pKa = 3.84FGVNAKK131 pKa = 7.62GTPKK135 pKa = 10.67YY136 pKa = 10.18STFCLLSNTLPPNSGVPPNHH156 pKa = 6.74RR157 pKa = 11.84LEE159 pKa = 4.42TTFFVYY165 pKa = 10.59LPVTSPGANNRR176 pKa = 11.84LIASFMAGFNLTTFFRR192 pKa = 11.84FCLRR196 pKa = 11.84TFPRR200 pKa = 11.84RR201 pKa = 11.84TLPAA205 pKa = 4.75
Molecular weight: 22.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
913 |
205 |
439 |
304.3 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.791 ± 0.4 | 0.548 ± 0.316 |
5.257 ± 1.517 | 4.491 ± 0.213 |
5.257 ± 1.682 | 8.653 ± 1.652 |
1.752 ± 0.377 | 4.381 ± 0.595 |
5.696 ± 0.953 | 8.105 ± 1.509 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.368 | 4.272 ± 1.045 |
5.696 ± 0.903 | 3.286 ± 0.689 |
7.338 ± 0.816 | 7.558 ± 0.933 |
6.243 ± 1.535 | 6.462 ± 0.811 |
2.3 ± 0.861 | 3.943 ± 0.709 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
