
Streptococcus phage SW31
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Brussowvirus; unclassified Brussowvirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
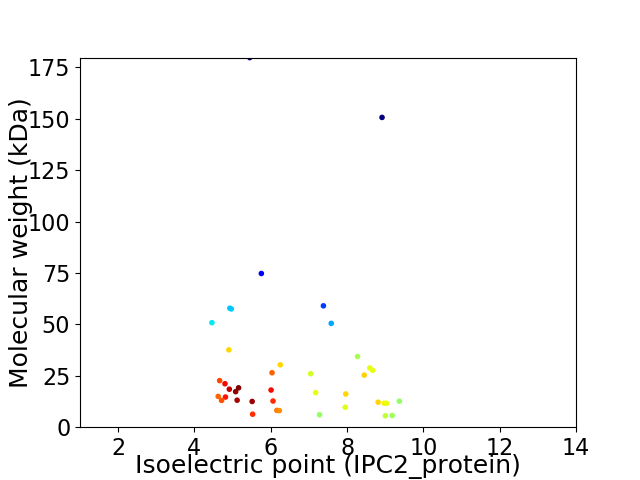
Virtual 2D-PAGE plot for 41 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A451EGR7|A0A451EGR7_9CAUD XRE family transcriptional regulator OS=Streptococcus phage SW31 OX=2419651 GN=SW31_028 PE=4 SV=1
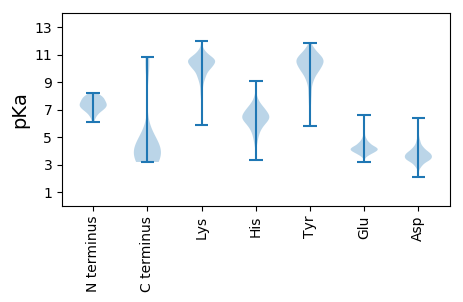
MM1 pKa = 6.11TTKK4 pKa = 9.29TQLLNTLDD12 pKa = 3.64SLVNQRR18 pKa = 11.84VTVPTNPFGGQCIALIDD35 pKa = 4.58NVLQYY40 pKa = 10.89QGLFNLDD47 pKa = 3.63FSYY50 pKa = 11.48LNAIDD55 pKa = 3.89ALSRR59 pKa = 11.84AEE61 pKa = 4.04SLGLKK66 pKa = 6.96VTRR69 pKa = 11.84FNGANNPTVGSVWVTNCLPYY89 pKa = 10.51HH90 pKa = 5.36QFGHH94 pKa = 6.24IGFVYY99 pKa = 10.67AEE101 pKa = 4.39NPDD104 pKa = 3.63GTVTTIEE111 pKa = 3.95QNIDD115 pKa = 3.02GNADD119 pKa = 3.22SLYY122 pKa = 10.62NGGWTRR128 pKa = 11.84KK129 pKa = 5.87VTRR132 pKa = 11.84NLDD135 pKa = 3.39SAGNFSYY142 pKa = 10.72IDD144 pKa = 3.43WNAPTQQMVGWFEE157 pKa = 4.33LPLDD161 pKa = 4.01GMEE164 pKa = 4.43KK165 pKa = 10.65DD166 pKa = 3.73NYY168 pKa = 10.34FIDD171 pKa = 3.66VSAYY175 pKa = 9.95QPGDD179 pKa = 3.26LTAICQASGTNNTVIKK195 pKa = 9.37VTEE198 pKa = 4.14GVGWVSPVAAQQTNTSNCIGYY219 pKa = 7.92YY220 pKa = 9.85HH221 pKa = 7.1FARR224 pKa = 11.84FGGDD228 pKa = 2.81VATAQAEE235 pKa = 4.18ANYY238 pKa = 9.8FISNLPSHH246 pKa = 6.82PRR248 pKa = 11.84YY249 pKa = 9.38LVCDD253 pKa = 4.05YY254 pKa = 11.33EE255 pKa = 6.63DD256 pKa = 4.21GASSDD261 pKa = 3.44KK262 pKa = 10.65QANTNAVLAFMDD274 pKa = 3.3ICKK277 pKa = 10.71ANGFEE282 pKa = 4.51PIYY285 pKa = 10.66YY286 pKa = 9.81SYY288 pKa = 11.36KK289 pKa = 10.34PYY291 pKa = 10.33TLANVYY297 pKa = 9.91VDD299 pKa = 4.33QITARR304 pKa = 11.84YY305 pKa = 7.8PNSLWIAAYY314 pKa = 8.94PDD316 pKa = 3.74YY317 pKa = 10.64EE318 pKa = 4.52VRR320 pKa = 11.84PEE322 pKa = 4.38PYY324 pKa = 9.07WGVYY328 pKa = 10.28PNMEE332 pKa = 4.23HH333 pKa = 5.47TRR335 pKa = 11.84WWQFTSTGLAGGLDD349 pKa = 3.79KK350 pKa = 11.4NIVIINDD357 pKa = 3.44DD358 pKa = 4.28DD359 pKa = 4.33NLVNQKK365 pKa = 9.68EE366 pKa = 4.45EE367 pKa = 3.98EE368 pKa = 4.64DD369 pKa = 3.22IMNFVVRR376 pKa = 11.84SEE378 pKa = 4.14SGKK381 pKa = 9.02EE382 pKa = 3.54GWVAVVNGRR391 pKa = 11.84VFGIGSMGTVDD402 pKa = 4.02ALEE405 pKa = 4.13ATGAKK410 pKa = 9.84RR411 pKa = 11.84LQLEE415 pKa = 4.23DD416 pKa = 3.99ADD418 pKa = 4.51FNRR421 pKa = 11.84FLYY424 pKa = 10.37SQSNDD429 pKa = 2.85TAAVAKK435 pKa = 10.67AIDD438 pKa = 3.8EE439 pKa = 4.55ASASVVKK446 pKa = 10.68AIEE449 pKa = 3.89EE450 pKa = 4.05RR451 pKa = 11.84AQATQGQTGKK461 pKa = 10.76
MM1 pKa = 6.11TTKK4 pKa = 9.29TQLLNTLDD12 pKa = 3.64SLVNQRR18 pKa = 11.84VTVPTNPFGGQCIALIDD35 pKa = 4.58NVLQYY40 pKa = 10.89QGLFNLDD47 pKa = 3.63FSYY50 pKa = 11.48LNAIDD55 pKa = 3.89ALSRR59 pKa = 11.84AEE61 pKa = 4.04SLGLKK66 pKa = 6.96VTRR69 pKa = 11.84FNGANNPTVGSVWVTNCLPYY89 pKa = 10.51HH90 pKa = 5.36QFGHH94 pKa = 6.24IGFVYY99 pKa = 10.67AEE101 pKa = 4.39NPDD104 pKa = 3.63GTVTTIEE111 pKa = 3.95QNIDD115 pKa = 3.02GNADD119 pKa = 3.22SLYY122 pKa = 10.62NGGWTRR128 pKa = 11.84KK129 pKa = 5.87VTRR132 pKa = 11.84NLDD135 pKa = 3.39SAGNFSYY142 pKa = 10.72IDD144 pKa = 3.43WNAPTQQMVGWFEE157 pKa = 4.33LPLDD161 pKa = 4.01GMEE164 pKa = 4.43KK165 pKa = 10.65DD166 pKa = 3.73NYY168 pKa = 10.34FIDD171 pKa = 3.66VSAYY175 pKa = 9.95QPGDD179 pKa = 3.26LTAICQASGTNNTVIKK195 pKa = 9.37VTEE198 pKa = 4.14GVGWVSPVAAQQTNTSNCIGYY219 pKa = 7.92YY220 pKa = 9.85HH221 pKa = 7.1FARR224 pKa = 11.84FGGDD228 pKa = 2.81VATAQAEE235 pKa = 4.18ANYY238 pKa = 9.8FISNLPSHH246 pKa = 6.82PRR248 pKa = 11.84YY249 pKa = 9.38LVCDD253 pKa = 4.05YY254 pKa = 11.33EE255 pKa = 6.63DD256 pKa = 4.21GASSDD261 pKa = 3.44KK262 pKa = 10.65QANTNAVLAFMDD274 pKa = 3.3ICKK277 pKa = 10.71ANGFEE282 pKa = 4.51PIYY285 pKa = 10.66YY286 pKa = 9.81SYY288 pKa = 11.36KK289 pKa = 10.34PYY291 pKa = 10.33TLANVYY297 pKa = 9.91VDD299 pKa = 4.33QITARR304 pKa = 11.84YY305 pKa = 7.8PNSLWIAAYY314 pKa = 8.94PDD316 pKa = 3.74YY317 pKa = 10.64EE318 pKa = 4.52VRR320 pKa = 11.84PEE322 pKa = 4.38PYY324 pKa = 9.07WGVYY328 pKa = 10.28PNMEE332 pKa = 4.23HH333 pKa = 5.47TRR335 pKa = 11.84WWQFTSTGLAGGLDD349 pKa = 3.79KK350 pKa = 11.4NIVIINDD357 pKa = 3.44DD358 pKa = 4.28DD359 pKa = 4.33NLVNQKK365 pKa = 9.68EE366 pKa = 4.45EE367 pKa = 3.98EE368 pKa = 4.64DD369 pKa = 3.22IMNFVVRR376 pKa = 11.84SEE378 pKa = 4.14SGKK381 pKa = 9.02EE382 pKa = 3.54GWVAVVNGRR391 pKa = 11.84VFGIGSMGTVDD402 pKa = 4.02ALEE405 pKa = 4.13ATGAKK410 pKa = 9.84RR411 pKa = 11.84LQLEE415 pKa = 4.23DD416 pKa = 3.99ADD418 pKa = 4.51FNRR421 pKa = 11.84FLYY424 pKa = 10.37SQSNDD429 pKa = 2.85TAAVAKK435 pKa = 10.67AIDD438 pKa = 3.8EE439 pKa = 4.55ASASVVKK446 pKa = 10.68AIEE449 pKa = 3.89EE450 pKa = 4.05RR451 pKa = 11.84AQATQGQTGKK461 pKa = 10.76
Molecular weight: 50.84 kDa
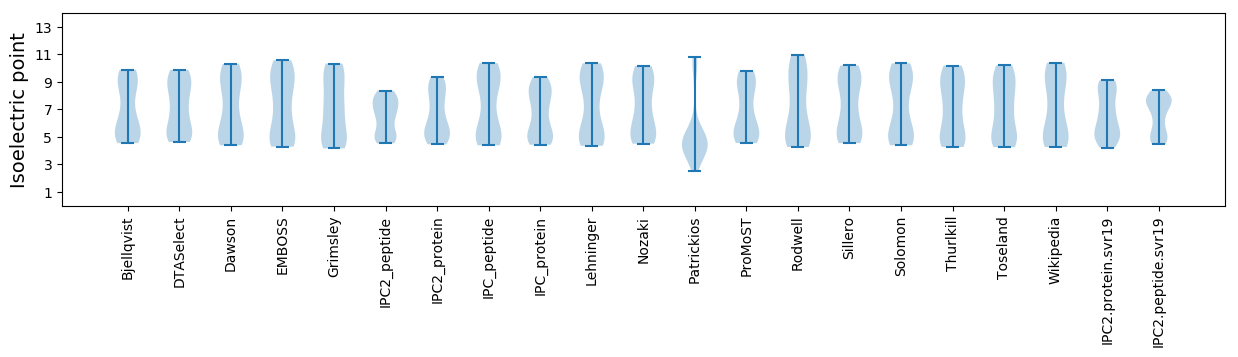
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S5X4M6|A0A3S5X4M6_9CAUD Uncharacterized protein OS=Streptococcus phage SW31 OX=2419651 GN=SW31_023 PE=4 SV=1
MM1 pKa = 7.83ADD3 pKa = 3.75FGVSLLEE10 pKa = 4.01ARR12 pKa = 11.84RR13 pKa = 11.84MTLKK17 pKa = 10.49EE18 pKa = 3.53MKK20 pKa = 9.92LYY22 pKa = 10.32QKK24 pKa = 10.55AYY26 pKa = 9.84KK27 pKa = 10.03KK28 pKa = 10.52RR29 pKa = 11.84FLNKK33 pKa = 9.43EE34 pKa = 3.69RR35 pKa = 11.84EE36 pKa = 4.36IYY38 pKa = 9.68QLAYY42 pKa = 10.87LNRR45 pKa = 11.84LANATTKK52 pKa = 10.62DD53 pKa = 3.03GKK55 pKa = 10.64KK56 pKa = 10.46YY57 pKa = 10.49YY58 pKa = 10.19FEE60 pKa = 5.48KK61 pKa = 10.83FDD63 pKa = 3.95DD64 pKa = 4.49FYY66 pKa = 11.56NAKK69 pKa = 9.54EE70 pKa = 3.83RR71 pKa = 11.84ARR73 pKa = 11.84EE74 pKa = 3.84VLGEE78 pKa = 4.35KK79 pKa = 8.61ITNSKK84 pKa = 10.18LLEE87 pKa = 3.98RR88 pKa = 11.84ARR90 pKa = 11.84NNLNYY95 pKa = 10.06KK96 pKa = 9.85KK97 pKa = 10.46EE98 pKa = 3.94RR99 pKa = 11.84GLLDD103 pKa = 3.22GRR105 pKa = 4.46
MM1 pKa = 7.83ADD3 pKa = 3.75FGVSLLEE10 pKa = 4.01ARR12 pKa = 11.84RR13 pKa = 11.84MTLKK17 pKa = 10.49EE18 pKa = 3.53MKK20 pKa = 9.92LYY22 pKa = 10.32QKK24 pKa = 10.55AYY26 pKa = 9.84KK27 pKa = 10.03KK28 pKa = 10.52RR29 pKa = 11.84FLNKK33 pKa = 9.43EE34 pKa = 3.69RR35 pKa = 11.84EE36 pKa = 4.36IYY38 pKa = 9.68QLAYY42 pKa = 10.87LNRR45 pKa = 11.84LANATTKK52 pKa = 10.62DD53 pKa = 3.03GKK55 pKa = 10.64KK56 pKa = 10.46YY57 pKa = 10.49YY58 pKa = 10.19FEE60 pKa = 5.48KK61 pKa = 10.83FDD63 pKa = 3.95DD64 pKa = 4.49FYY66 pKa = 11.56NAKK69 pKa = 9.54EE70 pKa = 3.83RR71 pKa = 11.84ARR73 pKa = 11.84EE74 pKa = 3.84VLGEE78 pKa = 4.35KK79 pKa = 8.61ITNSKK84 pKa = 10.18LLEE87 pKa = 3.98RR88 pKa = 11.84ARR90 pKa = 11.84NNLNYY95 pKa = 10.06KK96 pKa = 9.85KK97 pKa = 10.46EE98 pKa = 3.94RR99 pKa = 11.84GLLDD103 pKa = 3.22GRR105 pKa = 4.46
Molecular weight: 12.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11139 |
48 |
1609 |
271.7 |
30.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.685 ± 0.955 | 0.44 ± 0.112 |
5.97 ± 0.438 | 6.715 ± 0.678 |
3.932 ± 0.176 | 6.859 ± 0.528 |
1.23 ± 0.164 | 6.724 ± 0.362 |
7.721 ± 0.552 | 7.909 ± 0.377 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.343 ± 0.358 | 5.853 ± 0.373 |
3.052 ± 0.242 | 3.959 ± 0.24 |
4.273 ± 0.393 | 6.913 ± 0.653 |
6.616 ± 0.332 | 6.625 ± 0.201 |
1.329 ± 0.209 | 3.851 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
