
Nocardiopsaceae bacterium YIM 96095
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; unclassified Nocardiopsaceae
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
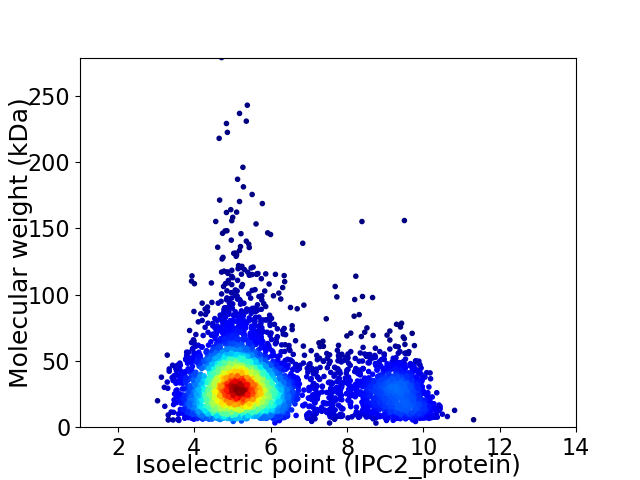
Virtual 2D-PAGE plot for 4542 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N0DQW5|A0A3N0DQW5_9ACTN O-phosphoserine phosphohydrolase OS=Nocardiopsaceae bacterium YIM 96095 OX=2487137 GN=serB PE=3 SV=1
MM1 pKa = 7.24IVSAVVAAVLMAGVPQRR18 pKa = 11.84IDD20 pKa = 3.58DD21 pKa = 5.51LITSGLARR29 pKa = 11.84ALGPSDD35 pKa = 4.41PGTGSNADD43 pKa = 3.55PGEE46 pKa = 4.25QYY48 pKa = 10.44WNQSDD53 pKa = 4.45PDD55 pKa = 4.01SSPATEE61 pKa = 4.68PEE63 pKa = 4.25HH64 pKa = 6.12VTDD67 pKa = 5.11PNHH70 pKa = 7.18PDD72 pKa = 3.52YY73 pKa = 10.95EE74 pKa = 4.66GPPAGHH80 pKa = 7.39PAGPPAPSNAGPDD93 pKa = 3.62PADD96 pKa = 4.01ADD98 pKa = 4.07SEE100 pKa = 4.37AVEE103 pKa = 4.07AAVEE107 pKa = 3.98EE108 pKa = 4.67LEE110 pKa = 5.04GCLSDD115 pKa = 3.85WNEE118 pKa = 3.83DD119 pKa = 3.78APWFTGTCAYY129 pKa = 7.9DD130 pKa = 3.47TYY132 pKa = 11.59KK133 pKa = 10.9DD134 pKa = 3.79LSPEE138 pKa = 3.73EE139 pKa = 3.91LAAVVDD145 pKa = 4.28TMSSQDD151 pKa = 3.47LGRR154 pKa = 11.84LFEE157 pKa = 4.82SGPIPLEE164 pKa = 3.52QEE166 pKa = 4.0YY167 pKa = 10.7FDD169 pKa = 4.58LVEE172 pKa = 4.82EE173 pKa = 4.39IQRR176 pKa = 11.84TAPLDD181 pKa = 3.52TLRR184 pKa = 11.84ALQNSGATPFAQPDD198 pKa = 3.87FEE200 pKa = 5.24GVGGDD205 pKa = 3.58PAGGNSPSNVSYY217 pKa = 11.65GEE219 pKa = 3.67IDD221 pKa = 4.06DD222 pKa = 3.92YY223 pKa = 11.93SLYY226 pKa = 11.26GEE228 pKa = 4.44GDD230 pKa = 3.66EE231 pKa = 4.27PVSWEE236 pKa = 4.1HH237 pKa = 6.44VNQNSLGDD245 pKa = 4.0CWLMATMGAMARR257 pKa = 11.84QNPGHH262 pKa = 5.9VEE264 pKa = 3.8EE265 pKa = 5.29MIQEE269 pKa = 4.33NANGTYY275 pKa = 9.81TVTFPGKK282 pKa = 10.34DD283 pKa = 3.45PVTVTPDD290 pKa = 3.2LPLNPDD296 pKa = 3.5GTPAFAGSNEE306 pKa = 4.36DD307 pKa = 4.8PPVIWPAIVEE317 pKa = 4.03KK318 pKa = 10.52AYY320 pKa = 10.77ADD322 pKa = 4.17LEE324 pKa = 4.53GNDD327 pKa = 3.71YY328 pKa = 11.62SNLEE332 pKa = 3.91NWHH335 pKa = 6.74AGAAMNTFAGGAGMDD350 pKa = 4.4YY351 pKa = 10.7IPDD354 pKa = 3.77VTDD357 pKa = 3.11TTMDD361 pKa = 3.33EE362 pKa = 4.17LAGKK366 pKa = 9.64FEE368 pKa = 4.43NGEE371 pKa = 4.64AITLSTPPTMQLNDD385 pKa = 3.44WPKK388 pKa = 11.0DD389 pKa = 3.57GTGKK393 pKa = 10.94GDD395 pKa = 4.92LVDD398 pKa = 5.67DD399 pKa = 4.68DD400 pKa = 5.67AGTLVSGHH408 pKa = 5.82VYY410 pKa = 10.76FMTGVNRR417 pKa = 11.84DD418 pKa = 3.79DD419 pKa = 3.75GTVTLRR425 pKa = 11.84NPWGSHH431 pKa = 4.63TDD433 pKa = 3.95DD434 pKa = 3.69IVLSLDD440 pKa = 3.48EE441 pKa = 4.63VNDD444 pKa = 3.35NFAAIHH450 pKa = 5.32STNLDD455 pKa = 3.47EE456 pKa = 5.24
MM1 pKa = 7.24IVSAVVAAVLMAGVPQRR18 pKa = 11.84IDD20 pKa = 3.58DD21 pKa = 5.51LITSGLARR29 pKa = 11.84ALGPSDD35 pKa = 4.41PGTGSNADD43 pKa = 3.55PGEE46 pKa = 4.25QYY48 pKa = 10.44WNQSDD53 pKa = 4.45PDD55 pKa = 4.01SSPATEE61 pKa = 4.68PEE63 pKa = 4.25HH64 pKa = 6.12VTDD67 pKa = 5.11PNHH70 pKa = 7.18PDD72 pKa = 3.52YY73 pKa = 10.95EE74 pKa = 4.66GPPAGHH80 pKa = 7.39PAGPPAPSNAGPDD93 pKa = 3.62PADD96 pKa = 4.01ADD98 pKa = 4.07SEE100 pKa = 4.37AVEE103 pKa = 4.07AAVEE107 pKa = 3.98EE108 pKa = 4.67LEE110 pKa = 5.04GCLSDD115 pKa = 3.85WNEE118 pKa = 3.83DD119 pKa = 3.78APWFTGTCAYY129 pKa = 7.9DD130 pKa = 3.47TYY132 pKa = 11.59KK133 pKa = 10.9DD134 pKa = 3.79LSPEE138 pKa = 3.73EE139 pKa = 3.91LAAVVDD145 pKa = 4.28TMSSQDD151 pKa = 3.47LGRR154 pKa = 11.84LFEE157 pKa = 4.82SGPIPLEE164 pKa = 3.52QEE166 pKa = 4.0YY167 pKa = 10.7FDD169 pKa = 4.58LVEE172 pKa = 4.82EE173 pKa = 4.39IQRR176 pKa = 11.84TAPLDD181 pKa = 3.52TLRR184 pKa = 11.84ALQNSGATPFAQPDD198 pKa = 3.87FEE200 pKa = 5.24GVGGDD205 pKa = 3.58PAGGNSPSNVSYY217 pKa = 11.65GEE219 pKa = 3.67IDD221 pKa = 4.06DD222 pKa = 3.92YY223 pKa = 11.93SLYY226 pKa = 11.26GEE228 pKa = 4.44GDD230 pKa = 3.66EE231 pKa = 4.27PVSWEE236 pKa = 4.1HH237 pKa = 6.44VNQNSLGDD245 pKa = 4.0CWLMATMGAMARR257 pKa = 11.84QNPGHH262 pKa = 5.9VEE264 pKa = 3.8EE265 pKa = 5.29MIQEE269 pKa = 4.33NANGTYY275 pKa = 9.81TVTFPGKK282 pKa = 10.34DD283 pKa = 3.45PVTVTPDD290 pKa = 3.2LPLNPDD296 pKa = 3.5GTPAFAGSNEE306 pKa = 4.36DD307 pKa = 4.8PPVIWPAIVEE317 pKa = 4.03KK318 pKa = 10.52AYY320 pKa = 10.77ADD322 pKa = 4.17LEE324 pKa = 4.53GNDD327 pKa = 3.71YY328 pKa = 11.62SNLEE332 pKa = 3.91NWHH335 pKa = 6.74AGAAMNTFAGGAGMDD350 pKa = 4.4YY351 pKa = 10.7IPDD354 pKa = 3.77VTDD357 pKa = 3.11TTMDD361 pKa = 3.33EE362 pKa = 4.17LAGKK366 pKa = 9.64FEE368 pKa = 4.43NGEE371 pKa = 4.64AITLSTPPTMQLNDD385 pKa = 3.44WPKK388 pKa = 11.0DD389 pKa = 3.57GTGKK393 pKa = 10.94GDD395 pKa = 4.92LVDD398 pKa = 5.67DD399 pKa = 4.68DD400 pKa = 5.67AGTLVSGHH408 pKa = 5.82VYY410 pKa = 10.76FMTGVNRR417 pKa = 11.84DD418 pKa = 3.79DD419 pKa = 3.75GTVTLRR425 pKa = 11.84NPWGSHH431 pKa = 4.63TDD433 pKa = 3.95DD434 pKa = 3.69IVLSLDD440 pKa = 3.48EE441 pKa = 4.63VNDD444 pKa = 3.35NFAAIHH450 pKa = 5.32STNLDD455 pKa = 3.47EE456 pKa = 5.24
Molecular weight: 48.36 kDa
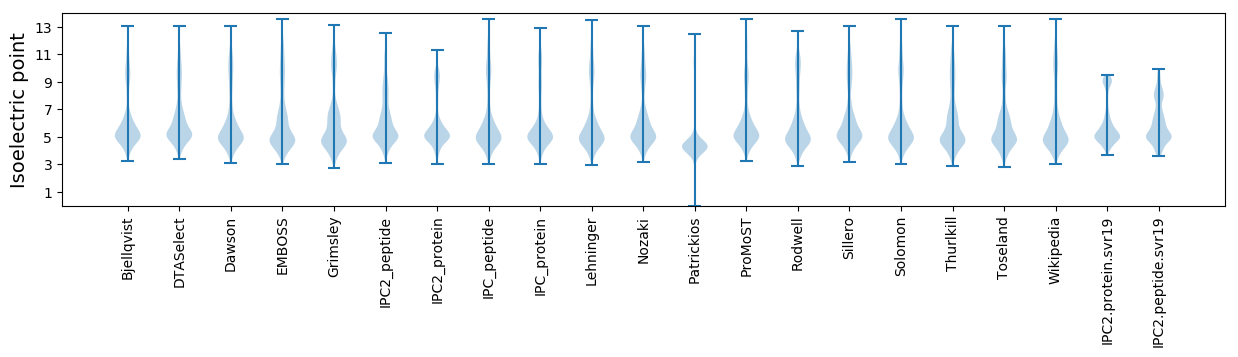
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N0E5S4|A0A3N0E5S4_9ACTN DUF721 domain-containing protein OS=Nocardiopsaceae bacterium YIM 96095 OX=2487137 GN=EFW17_16885 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.68KK38 pKa = 9.79GRR40 pKa = 11.84KK41 pKa = 8.94RR42 pKa = 11.84LTVSHH47 pKa = 6.96
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.68KK38 pKa = 9.79GRR40 pKa = 11.84KK41 pKa = 8.94RR42 pKa = 11.84LTVSHH47 pKa = 6.96
Molecular weight: 5.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1459053 |
29 |
2590 |
321.2 |
34.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.302 ± 0.05 | 0.762 ± 0.01 |
6.247 ± 0.035 | 6.765 ± 0.041 |
2.752 ± 0.021 | 9.198 ± 0.034 |
2.404 ± 0.02 | 3.366 ± 0.024 |
1.513 ± 0.023 | 10.116 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.902 ± 0.015 | 1.893 ± 0.016 |
5.939 ± 0.034 | 2.554 ± 0.02 |
8.386 ± 0.038 | 5.421 ± 0.025 |
6.17 ± 0.024 | 8.781 ± 0.038 |
1.509 ± 0.014 | 2.019 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
