
Pontiella sulfatireligans
Taxonomy: cellular organisms; Bacteria; PVC group; Kiritimatiellaeota; Kiritimatiellae; Kiritimatiellales; Pontiellaceae; Pontiella
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
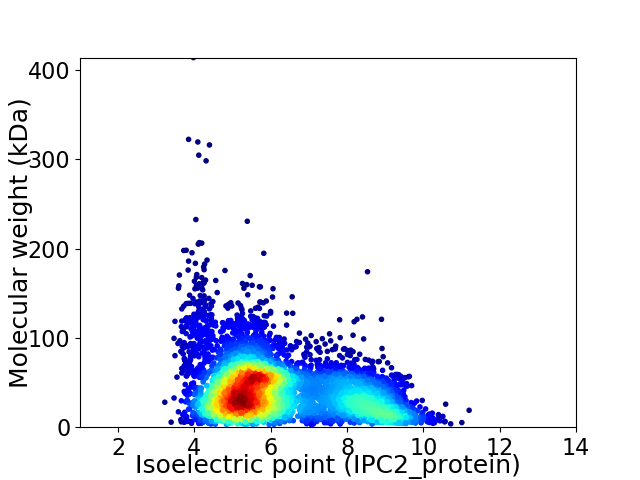
Virtual 2D-PAGE plot for 5575 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C2UL90|A0A6C2UL90_9BACT Uncharacterized protein OS=Pontiella sulfatireligans OX=2750658 GN=SCARR_03069 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 9.86MRR4 pKa = 11.84GITLFAALLGMAIGTQAALPEE25 pKa = 4.61IGDD28 pKa = 4.36TIWLKK33 pKa = 11.17GGTDD37 pKa = 3.15NSYY40 pKa = 11.26VYY42 pKa = 10.89ASGSALVAKK51 pKa = 10.75AEE53 pKa = 4.12TDD55 pKa = 3.45VANALNFVVHH65 pKa = 6.32SADD68 pKa = 3.55GSDD71 pKa = 3.44SNFIFQVADD80 pKa = 3.38TTSYY84 pKa = 11.24VIVVTNNDD92 pKa = 4.37DD93 pKa = 3.56ILTIDD98 pKa = 3.9ATALTNNPLTHH109 pKa = 6.23FTMTEE114 pKa = 3.75ITEE117 pKa = 3.94GTYY120 pKa = 10.73AGTYY124 pKa = 8.86EE125 pKa = 4.53LSCLAHH131 pKa = 7.11ADD133 pKa = 3.81PVVHH137 pKa = 6.48EE138 pKa = 4.99RR139 pKa = 11.84GNTHH143 pKa = 5.5TLKK146 pKa = 10.8AHH148 pKa = 5.12STSGGSIRR156 pKa = 11.84SAWTYY161 pKa = 10.51GVVGAALEE169 pKa = 4.4PPTGLGANTSDD180 pKa = 3.62SQVVLDD186 pKa = 4.11WDD188 pKa = 3.93NDD190 pKa = 3.56VSGLLDD196 pKa = 3.62TYY198 pKa = 9.92TVYY201 pKa = 10.98RR202 pKa = 11.84STNTPVTISDD212 pKa = 3.95TLLGSPSNSQFTDD225 pKa = 3.41TNVVNGTTYY234 pKa = 10.8YY235 pKa = 10.52YY236 pKa = 11.19AVTATDD242 pKa = 4.35GSTEE246 pKa = 3.93TDD248 pKa = 3.11LSNEE252 pKa = 3.96VSATPPTGTAQPKK265 pKa = 9.13RR266 pKa = 11.84LKK268 pKa = 10.89ALGVPAAVLLDD279 pKa = 3.59WADD282 pKa = 3.63DD283 pKa = 4.05TTGHH287 pKa = 6.94LDD289 pKa = 3.25YY290 pKa = 10.29YY291 pKa = 9.17TVYY294 pKa = 10.76RR295 pKa = 11.84STTTPVTTGSPVLTNVTASAFTDD318 pKa = 3.99FDD320 pKa = 4.11VVAGTTNYY328 pKa = 10.46YY329 pKa = 11.0AVTAVGTNGVPPEE342 pKa = 4.22SALSDD347 pKa = 3.67EE348 pKa = 4.67VSAIPFAAVTEE359 pKa = 4.56TVLFQHH365 pKa = 6.95IDD367 pKa = 3.19ATVAASVQTDD377 pKa = 3.47GVTNGMGIVTDD388 pKa = 4.02VVDD391 pKa = 4.11QSTFGNDD398 pKa = 2.95ATDD401 pKa = 3.46VADD404 pKa = 5.2GDD406 pKa = 4.32DD407 pKa = 4.8LGPVLWPSTDD417 pKa = 3.13LFGSGRR423 pKa = 11.84AGFDD427 pKa = 2.72MGTDD431 pKa = 3.17MRR433 pKa = 11.84MLNLFTNGTDD443 pKa = 3.24ALLDD447 pKa = 3.79FTGDD451 pKa = 3.28ASGSDD456 pKa = 3.64GFAMLVAFKK465 pKa = 10.13TRR467 pKa = 11.84TMTEE471 pKa = 3.73GVANIFLNNGPISMEE486 pKa = 3.7YY487 pKa = 10.62DD488 pKa = 3.44GDD490 pKa = 4.16DD491 pKa = 3.67GEE493 pKa = 4.68MTVEE497 pKa = 4.54LGSATLTQSGAGVEE511 pKa = 4.21AGDD514 pKa = 4.0TIVYY518 pKa = 9.66ALNYY522 pKa = 9.29NALTGEE528 pKa = 4.26TTFWDD533 pKa = 3.6SKK535 pKa = 10.71NNEE538 pKa = 4.3EE539 pKa = 4.54VSVTAEE545 pKa = 3.64AHH547 pKa = 6.43GDD549 pKa = 3.65FSGDD553 pKa = 3.47PLQLGGSGRR562 pKa = 11.84DD563 pKa = 3.27SRR565 pKa = 11.84IFDD568 pKa = 3.5GLMGEE573 pKa = 4.38VKK575 pKa = 10.22IFSTKK580 pKa = 10.37LPADD584 pKa = 3.27QFEE587 pKa = 4.43AQRR590 pKa = 11.84DD591 pKa = 3.74ALRR594 pKa = 11.84LKK596 pKa = 9.46WVSADD601 pKa = 3.16ATGFDD606 pKa = 3.72VWVSSWGVNLGSATNDD622 pKa = 3.01WDD624 pKa = 3.87NDD626 pKa = 4.41GINNLAEE633 pKa = 3.98YY634 pKa = 10.53AFAGNPTNGTDD645 pKa = 3.45DD646 pKa = 3.96TYY648 pKa = 11.58TDD650 pKa = 3.97LVTDD654 pKa = 4.26GGLVYY659 pKa = 10.49IHH661 pKa = 6.67PQPKK665 pKa = 9.78NDD667 pKa = 3.6ADD669 pKa = 3.88LNYY672 pKa = 10.99AVLTMDD678 pKa = 4.51NLPYY682 pKa = 9.81DD683 pKa = 3.26TWTDD687 pKa = 3.06EE688 pKa = 4.46GYY690 pKa = 8.14TAVIGTYY697 pKa = 10.04VPAEE701 pKa = 3.55GDD703 pKa = 3.5YY704 pKa = 11.17NYY706 pKa = 10.5VSNSVPTTDD715 pKa = 2.67AVKK718 pKa = 10.31FIRR721 pKa = 11.84ASAEE725 pKa = 3.91YY726 pKa = 10.63
MM1 pKa = 7.63KK2 pKa = 9.86MRR4 pKa = 11.84GITLFAALLGMAIGTQAALPEE25 pKa = 4.61IGDD28 pKa = 4.36TIWLKK33 pKa = 11.17GGTDD37 pKa = 3.15NSYY40 pKa = 11.26VYY42 pKa = 10.89ASGSALVAKK51 pKa = 10.75AEE53 pKa = 4.12TDD55 pKa = 3.45VANALNFVVHH65 pKa = 6.32SADD68 pKa = 3.55GSDD71 pKa = 3.44SNFIFQVADD80 pKa = 3.38TTSYY84 pKa = 11.24VIVVTNNDD92 pKa = 4.37DD93 pKa = 3.56ILTIDD98 pKa = 3.9ATALTNNPLTHH109 pKa = 6.23FTMTEE114 pKa = 3.75ITEE117 pKa = 3.94GTYY120 pKa = 10.73AGTYY124 pKa = 8.86EE125 pKa = 4.53LSCLAHH131 pKa = 7.11ADD133 pKa = 3.81PVVHH137 pKa = 6.48EE138 pKa = 4.99RR139 pKa = 11.84GNTHH143 pKa = 5.5TLKK146 pKa = 10.8AHH148 pKa = 5.12STSGGSIRR156 pKa = 11.84SAWTYY161 pKa = 10.51GVVGAALEE169 pKa = 4.4PPTGLGANTSDD180 pKa = 3.62SQVVLDD186 pKa = 4.11WDD188 pKa = 3.93NDD190 pKa = 3.56VSGLLDD196 pKa = 3.62TYY198 pKa = 9.92TVYY201 pKa = 10.98RR202 pKa = 11.84STNTPVTISDD212 pKa = 3.95TLLGSPSNSQFTDD225 pKa = 3.41TNVVNGTTYY234 pKa = 10.8YY235 pKa = 10.52YY236 pKa = 11.19AVTATDD242 pKa = 4.35GSTEE246 pKa = 3.93TDD248 pKa = 3.11LSNEE252 pKa = 3.96VSATPPTGTAQPKK265 pKa = 9.13RR266 pKa = 11.84LKK268 pKa = 10.89ALGVPAAVLLDD279 pKa = 3.59WADD282 pKa = 3.63DD283 pKa = 4.05TTGHH287 pKa = 6.94LDD289 pKa = 3.25YY290 pKa = 10.29YY291 pKa = 9.17TVYY294 pKa = 10.76RR295 pKa = 11.84STTTPVTTGSPVLTNVTASAFTDD318 pKa = 3.99FDD320 pKa = 4.11VVAGTTNYY328 pKa = 10.46YY329 pKa = 11.0AVTAVGTNGVPPEE342 pKa = 4.22SALSDD347 pKa = 3.67EE348 pKa = 4.67VSAIPFAAVTEE359 pKa = 4.56TVLFQHH365 pKa = 6.95IDD367 pKa = 3.19ATVAASVQTDD377 pKa = 3.47GVTNGMGIVTDD388 pKa = 4.02VVDD391 pKa = 4.11QSTFGNDD398 pKa = 2.95ATDD401 pKa = 3.46VADD404 pKa = 5.2GDD406 pKa = 4.32DD407 pKa = 4.8LGPVLWPSTDD417 pKa = 3.13LFGSGRR423 pKa = 11.84AGFDD427 pKa = 2.72MGTDD431 pKa = 3.17MRR433 pKa = 11.84MLNLFTNGTDD443 pKa = 3.24ALLDD447 pKa = 3.79FTGDD451 pKa = 3.28ASGSDD456 pKa = 3.64GFAMLVAFKK465 pKa = 10.13TRR467 pKa = 11.84TMTEE471 pKa = 3.73GVANIFLNNGPISMEE486 pKa = 3.7YY487 pKa = 10.62DD488 pKa = 3.44GDD490 pKa = 4.16DD491 pKa = 3.67GEE493 pKa = 4.68MTVEE497 pKa = 4.54LGSATLTQSGAGVEE511 pKa = 4.21AGDD514 pKa = 4.0TIVYY518 pKa = 9.66ALNYY522 pKa = 9.29NALTGEE528 pKa = 4.26TTFWDD533 pKa = 3.6SKK535 pKa = 10.71NNEE538 pKa = 4.3EE539 pKa = 4.54VSVTAEE545 pKa = 3.64AHH547 pKa = 6.43GDD549 pKa = 3.65FSGDD553 pKa = 3.47PLQLGGSGRR562 pKa = 11.84DD563 pKa = 3.27SRR565 pKa = 11.84IFDD568 pKa = 3.5GLMGEE573 pKa = 4.38VKK575 pKa = 10.22IFSTKK580 pKa = 10.37LPADD584 pKa = 3.27QFEE587 pKa = 4.43AQRR590 pKa = 11.84DD591 pKa = 3.74ALRR594 pKa = 11.84LKK596 pKa = 9.46WVSADD601 pKa = 3.16ATGFDD606 pKa = 3.72VWVSSWGVNLGSATNDD622 pKa = 3.01WDD624 pKa = 3.87NDD626 pKa = 4.41GINNLAEE633 pKa = 3.98YY634 pKa = 10.53AFAGNPTNGTDD645 pKa = 3.45DD646 pKa = 3.96TYY648 pKa = 11.58TDD650 pKa = 3.97LVTDD654 pKa = 4.26GGLVYY659 pKa = 10.49IHH661 pKa = 6.67PQPKK665 pKa = 9.78NDD667 pKa = 3.6ADD669 pKa = 3.88LNYY672 pKa = 10.99AVLTMDD678 pKa = 4.51NLPYY682 pKa = 9.81DD683 pKa = 3.26TWTDD687 pKa = 3.06EE688 pKa = 4.46GYY690 pKa = 8.14TAVIGTYY697 pKa = 10.04VPAEE701 pKa = 3.55GDD703 pKa = 3.5YY704 pKa = 11.17NYY706 pKa = 10.5VSNSVPTTDD715 pKa = 2.67AVKK718 pKa = 10.31FIRR721 pKa = 11.84ASAEE725 pKa = 3.91YY726 pKa = 10.63
Molecular weight: 76.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C2US34|A0A6C2US34_9BACT Diaminobutyrate--2-oxoglutarate transaminase OS=Pontiella sulfatireligans OX=2750658 GN=ectB PE=3 SV=1
MM1 pKa = 7.73KK2 pKa = 10.12KK3 pKa = 9.79RR4 pKa = 11.84KK5 pKa = 9.32KK6 pKa = 9.06KK7 pKa = 9.89HH8 pKa = 5.77RR9 pKa = 11.84PRR11 pKa = 11.84KK12 pKa = 9.55HH13 pKa = 5.39NQQHH17 pKa = 5.14TGKK20 pKa = 9.41VAYY23 pKa = 8.78MNSGRR28 pKa = 11.84IKK30 pKa = 10.08STLQTMRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MSS42 pKa = 2.94
MM1 pKa = 7.73KK2 pKa = 10.12KK3 pKa = 9.79RR4 pKa = 11.84KK5 pKa = 9.32KK6 pKa = 9.06KK7 pKa = 9.89HH8 pKa = 5.77RR9 pKa = 11.84PRR11 pKa = 11.84KK12 pKa = 9.55HH13 pKa = 5.39NQQHH17 pKa = 5.14TGKK20 pKa = 9.41VAYY23 pKa = 8.78MNSGRR28 pKa = 11.84IKK30 pKa = 10.08STLQTMRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MSS42 pKa = 2.94
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2209931 |
29 |
3911 |
396.4 |
43.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.858 ± 0.031 | 1.218 ± 0.012 |
6.067 ± 0.025 | 6.303 ± 0.03 |
4.17 ± 0.015 | 8.237 ± 0.031 |
2.174 ± 0.017 | 5.472 ± 0.022 |
5.227 ± 0.034 | 8.987 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.013 | 4.2 ± 0.024 |
4.694 ± 0.022 | 3.279 ± 0.017 |
4.949 ± 0.026 | 6.147 ± 0.027 |
5.578 ± 0.03 | 6.801 ± 0.028 |
1.679 ± 0.015 | 3.366 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
