
Sewage-associated circular DNA virus-5
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
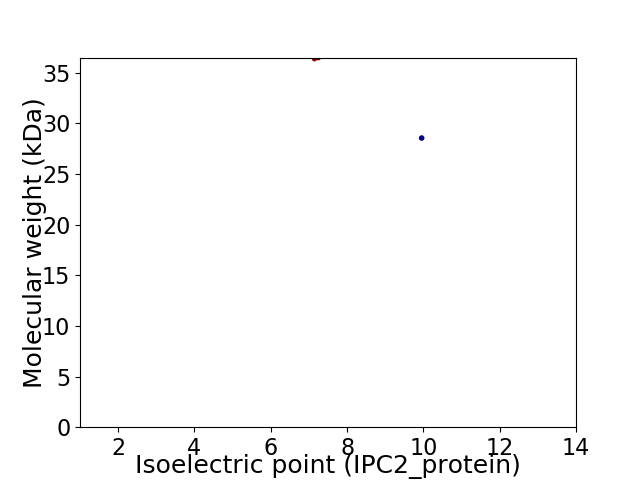
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IZU0|A0A075IZU0_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-5 OX=1519394 PE=4 SV=1
MM1 pKa = 8.01EE2 pKa = 5.24DD3 pKa = 3.22TLRR6 pKa = 11.84TPDD9 pKa = 3.67SEE11 pKa = 4.65TVKK14 pKa = 11.06AKK16 pKa = 10.4GDD18 pKa = 3.29QRR20 pKa = 11.84YY21 pKa = 9.77RR22 pKa = 11.84NVFYY26 pKa = 10.13TYY28 pKa = 11.26NNPTKK33 pKa = 10.29TDD35 pKa = 4.15EE36 pKa = 3.84EE37 pKa = 4.23WMDD40 pKa = 3.62YY41 pKa = 10.23VRR43 pKa = 11.84EE44 pKa = 3.96RR45 pKa = 11.84VPIAFHH51 pKa = 5.7VCALEE56 pKa = 4.4EE57 pKa = 4.51GDD59 pKa = 5.36LNHH62 pKa = 6.73TPHH65 pKa = 6.43LQGYY69 pKa = 7.46MEE71 pKa = 4.86FRR73 pKa = 11.84QQVRR77 pKa = 11.84KK78 pKa = 9.6SQLITAFGTHH88 pKa = 5.18FWFRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84GTALEE100 pKa = 4.1AYY102 pKa = 7.63EE103 pKa = 4.31YY104 pKa = 8.39CTKK107 pKa = 10.32SGKK110 pKa = 9.1YY111 pKa = 9.95AEE113 pKa = 4.91RR114 pKa = 11.84SHH116 pKa = 5.42EE117 pKa = 4.32TKK119 pKa = 10.59AGPWEE124 pKa = 4.0EE125 pKa = 4.52GKK127 pKa = 10.57RR128 pKa = 11.84SSPGHH133 pKa = 5.34RR134 pKa = 11.84TDD136 pKa = 3.81IEE138 pKa = 4.36AAVHH142 pKa = 5.74TLLKK146 pKa = 10.77DD147 pKa = 3.73GLQSAVTQHH156 pKa = 5.45TQTMVQWGKK165 pKa = 10.85GIKK168 pKa = 9.64QVVKK172 pKa = 10.73DD173 pKa = 4.03MRR175 pKa = 11.84DD176 pKa = 3.38PPPIEE181 pKa = 3.8RR182 pKa = 11.84TIEE185 pKa = 4.05VILLHH190 pKa = 6.67GPGGVGKK197 pKa = 9.74DD198 pKa = 3.13HH199 pKa = 5.81QVYY202 pKa = 8.01VTGEE206 pKa = 3.43KK207 pKa = 9.88SQRR210 pKa = 11.84TFHH213 pKa = 6.97LLPGDD218 pKa = 3.85LEE220 pKa = 4.11NRR222 pKa = 11.84TPYY225 pKa = 10.22EE226 pKa = 4.18GQSVLILDD234 pKa = 4.34EE235 pKa = 4.46FDD237 pKa = 4.06GASSRR242 pKa = 11.84ADD244 pKa = 3.07VRR246 pKa = 11.84TLNRR250 pKa = 11.84ILDD253 pKa = 3.53KK254 pKa = 10.61WPYY257 pKa = 9.64DD258 pKa = 3.84LKK260 pKa = 11.22GLYY263 pKa = 9.43TNEE266 pKa = 3.71PARR269 pKa = 11.84WTRR272 pKa = 11.84VYY274 pKa = 10.66ILTNSHH280 pKa = 5.87PLKK283 pKa = 10.25WYY285 pKa = 9.91DD286 pKa = 3.01WDD288 pKa = 4.19KK289 pKa = 11.52EE290 pKa = 4.42SLLSSHH296 pKa = 6.57TPNQQGVALEE306 pKa = 3.94PRR308 pKa = 11.84ARR310 pKa = 11.84SARR313 pKa = 11.84TRR315 pKa = 3.29
MM1 pKa = 8.01EE2 pKa = 5.24DD3 pKa = 3.22TLRR6 pKa = 11.84TPDD9 pKa = 3.67SEE11 pKa = 4.65TVKK14 pKa = 11.06AKK16 pKa = 10.4GDD18 pKa = 3.29QRR20 pKa = 11.84YY21 pKa = 9.77RR22 pKa = 11.84NVFYY26 pKa = 10.13TYY28 pKa = 11.26NNPTKK33 pKa = 10.29TDD35 pKa = 4.15EE36 pKa = 3.84EE37 pKa = 4.23WMDD40 pKa = 3.62YY41 pKa = 10.23VRR43 pKa = 11.84EE44 pKa = 3.96RR45 pKa = 11.84VPIAFHH51 pKa = 5.7VCALEE56 pKa = 4.4EE57 pKa = 4.51GDD59 pKa = 5.36LNHH62 pKa = 6.73TPHH65 pKa = 6.43LQGYY69 pKa = 7.46MEE71 pKa = 4.86FRR73 pKa = 11.84QQVRR77 pKa = 11.84KK78 pKa = 9.6SQLITAFGTHH88 pKa = 5.18FWFRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84GTALEE100 pKa = 4.1AYY102 pKa = 7.63EE103 pKa = 4.31YY104 pKa = 8.39CTKK107 pKa = 10.32SGKK110 pKa = 9.1YY111 pKa = 9.95AEE113 pKa = 4.91RR114 pKa = 11.84SHH116 pKa = 5.42EE117 pKa = 4.32TKK119 pKa = 10.59AGPWEE124 pKa = 4.0EE125 pKa = 4.52GKK127 pKa = 10.57RR128 pKa = 11.84SSPGHH133 pKa = 5.34RR134 pKa = 11.84TDD136 pKa = 3.81IEE138 pKa = 4.36AAVHH142 pKa = 5.74TLLKK146 pKa = 10.77DD147 pKa = 3.73GLQSAVTQHH156 pKa = 5.45TQTMVQWGKK165 pKa = 10.85GIKK168 pKa = 9.64QVVKK172 pKa = 10.73DD173 pKa = 4.03MRR175 pKa = 11.84DD176 pKa = 3.38PPPIEE181 pKa = 3.8RR182 pKa = 11.84TIEE185 pKa = 4.05VILLHH190 pKa = 6.67GPGGVGKK197 pKa = 9.74DD198 pKa = 3.13HH199 pKa = 5.81QVYY202 pKa = 8.01VTGEE206 pKa = 3.43KK207 pKa = 9.88SQRR210 pKa = 11.84TFHH213 pKa = 6.97LLPGDD218 pKa = 3.85LEE220 pKa = 4.11NRR222 pKa = 11.84TPYY225 pKa = 10.22EE226 pKa = 4.18GQSVLILDD234 pKa = 4.34EE235 pKa = 4.46FDD237 pKa = 4.06GASSRR242 pKa = 11.84ADD244 pKa = 3.07VRR246 pKa = 11.84TLNRR250 pKa = 11.84ILDD253 pKa = 3.53KK254 pKa = 10.61WPYY257 pKa = 9.64DD258 pKa = 3.84LKK260 pKa = 11.22GLYY263 pKa = 9.43TNEE266 pKa = 3.71PARR269 pKa = 11.84WTRR272 pKa = 11.84VYY274 pKa = 10.66ILTNSHH280 pKa = 5.87PLKK283 pKa = 10.25WYY285 pKa = 9.91DD286 pKa = 3.01WDD288 pKa = 4.19KK289 pKa = 11.52EE290 pKa = 4.42SLLSSHH296 pKa = 6.57TPNQQGVALEE306 pKa = 3.94PRR308 pKa = 11.84ARR310 pKa = 11.84SARR313 pKa = 11.84TRR315 pKa = 3.29
Molecular weight: 36.4 kDa
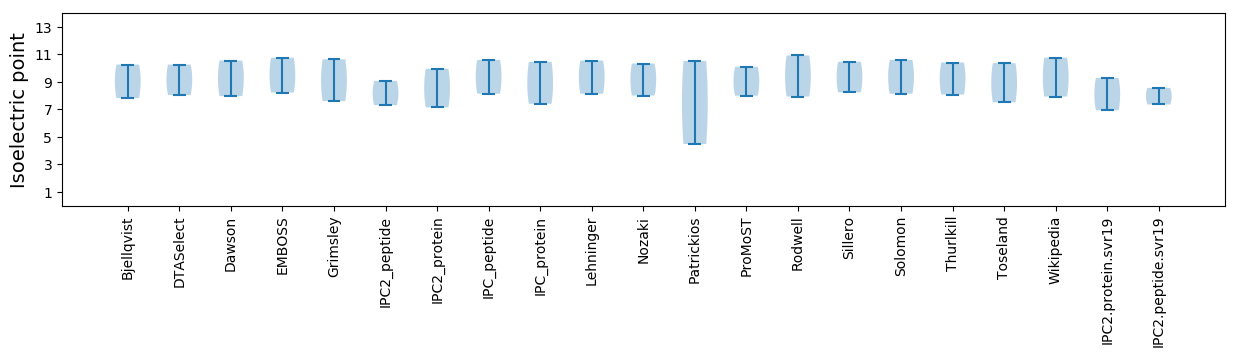
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075IZU0|A0A075IZU0_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-5 OX=1519394 PE=4 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84YY4 pKa = 6.71TKK6 pKa = 10.37KK7 pKa = 9.98SSSYY11 pKa = 9.2RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.96GVRR17 pKa = 11.84KK18 pKa = 7.26TRR20 pKa = 11.84AARR23 pKa = 11.84RR24 pKa = 11.84TYY26 pKa = 9.91RR27 pKa = 11.84KK28 pKa = 9.43KK29 pKa = 10.35GKK31 pKa = 9.53RR32 pKa = 11.84PSRR35 pKa = 11.84SRR37 pKa = 11.84IQTKK41 pKa = 10.34RR42 pKa = 11.84LGSRR46 pKa = 11.84YY47 pKa = 9.32PLGQKK52 pKa = 10.52AIIKK56 pKa = 10.07LNYY59 pKa = 8.68IGNGQVTGGGVLGYY73 pKa = 10.69GSGGTYY79 pKa = 9.26TLNSLSSPCKK89 pKa = 10.53NGTTDD94 pKa = 2.8NRR96 pKa = 11.84AAKK99 pKa = 9.62FLPNSFEE106 pKa = 4.36KK107 pKa = 11.06YY108 pKa = 9.13RR109 pKa = 11.84VTGMSYY115 pKa = 10.47RR116 pKa = 11.84IAATLVNTATPVPGYY131 pKa = 8.53MWFVPFEE138 pKa = 4.67AGTTTMPGTSAYY150 pKa = 10.24GDD152 pKa = 3.73QGEE155 pKa = 4.3MAQIPGAKK163 pKa = 9.5FKK165 pKa = 11.12NLEE168 pKa = 3.71AWNASGKK175 pKa = 8.41MAYY178 pKa = 10.15LKK180 pKa = 10.68GYY182 pKa = 11.21VNMKK186 pKa = 9.01QMRR189 pKa = 11.84GDD191 pKa = 3.44GRR193 pKa = 11.84SAGDD197 pKa = 3.77DD198 pKa = 3.75DD199 pKa = 4.09FTGTVGISGNWTDD212 pKa = 5.37PSRR215 pKa = 11.84LWDD218 pKa = 3.63YY219 pKa = 11.78AFGWSSINRR228 pKa = 11.84TPVPTTQAMHH238 pKa = 5.54YY239 pKa = 9.56SIRR242 pKa = 11.84TTYY245 pKa = 9.86YY246 pKa = 9.94VEE248 pKa = 4.3FFSPSMNGIEE258 pKa = 3.96
MM1 pKa = 7.51ARR3 pKa = 11.84YY4 pKa = 6.71TKK6 pKa = 10.37KK7 pKa = 9.98SSSYY11 pKa = 9.2RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.96GVRR17 pKa = 11.84KK18 pKa = 7.26TRR20 pKa = 11.84AARR23 pKa = 11.84RR24 pKa = 11.84TYY26 pKa = 9.91RR27 pKa = 11.84KK28 pKa = 9.43KK29 pKa = 10.35GKK31 pKa = 9.53RR32 pKa = 11.84PSRR35 pKa = 11.84SRR37 pKa = 11.84IQTKK41 pKa = 10.34RR42 pKa = 11.84LGSRR46 pKa = 11.84YY47 pKa = 9.32PLGQKK52 pKa = 10.52AIIKK56 pKa = 10.07LNYY59 pKa = 8.68IGNGQVTGGGVLGYY73 pKa = 10.69GSGGTYY79 pKa = 9.26TLNSLSSPCKK89 pKa = 10.53NGTTDD94 pKa = 2.8NRR96 pKa = 11.84AAKK99 pKa = 9.62FLPNSFEE106 pKa = 4.36KK107 pKa = 11.06YY108 pKa = 9.13RR109 pKa = 11.84VTGMSYY115 pKa = 10.47RR116 pKa = 11.84IAATLVNTATPVPGYY131 pKa = 8.53MWFVPFEE138 pKa = 4.67AGTTTMPGTSAYY150 pKa = 10.24GDD152 pKa = 3.73QGEE155 pKa = 4.3MAQIPGAKK163 pKa = 9.5FKK165 pKa = 11.12NLEE168 pKa = 3.71AWNASGKK175 pKa = 8.41MAYY178 pKa = 10.15LKK180 pKa = 10.68GYY182 pKa = 11.21VNMKK186 pKa = 9.01QMRR189 pKa = 11.84GDD191 pKa = 3.44GRR193 pKa = 11.84SAGDD197 pKa = 3.77DD198 pKa = 3.75DD199 pKa = 4.09FTGTVGISGNWTDD212 pKa = 5.37PSRR215 pKa = 11.84LWDD218 pKa = 3.63YY219 pKa = 11.78AFGWSSINRR228 pKa = 11.84TPVPTTQAMHH238 pKa = 5.54YY239 pKa = 9.56SIRR242 pKa = 11.84TTYY245 pKa = 9.86YY246 pKa = 9.94VEE248 pKa = 4.3FFSPSMNGIEE258 pKa = 3.96
Molecular weight: 28.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
573 |
258 |
315 |
286.5 |
32.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.283 ± 0.633 | 0.524 ± 0.08 |
4.712 ± 0.944 | 5.236 ± 1.704 |
2.967 ± 0.305 | 9.25 ± 1.62 |
2.443 ± 1.204 | 3.49 ± 0.226 |
6.283 ± 0.406 | 6.283 ± 1.183 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.737 | 3.839 ± 0.702 |
5.41 ± 0.218 | 3.839 ± 0.66 |
8.202 ± 0.037 | 6.632 ± 1.11 |
9.075 ± 0.36 | 5.236 ± 0.569 |
2.269 ± 0.194 | 5.41 ± 0.691 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
