
Marchantia polymorpha subsp. ruderalis
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia; Marchantia polymorpha
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
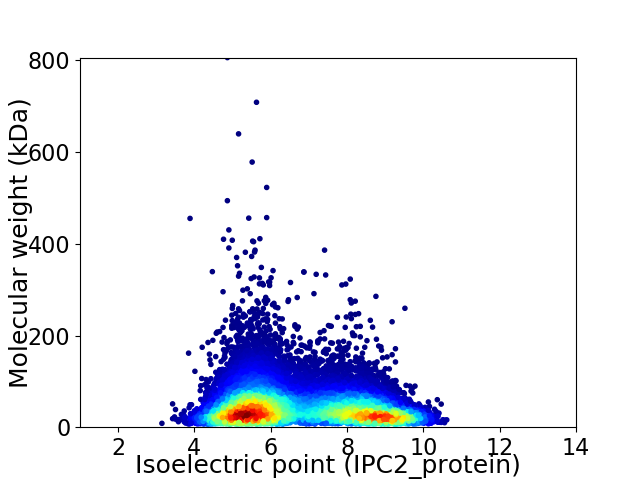
Virtual 2D-PAGE plot for 17951 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A176VNU6|A0A176VNU6_MARPO Uncharacterized protein OS=Marchantia polymorpha subsp. ruderalis OX=1480154 GN=AXG93_242s1340 PE=4 SV=1
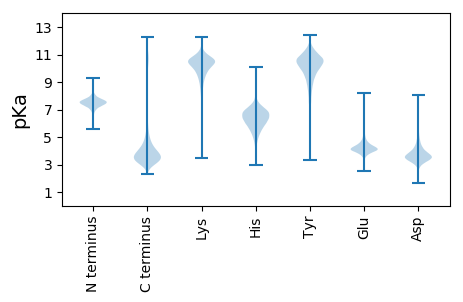
MM1 pKa = 7.39AQTEE5 pKa = 4.19LALPSGRR12 pKa = 11.84LQRR15 pKa = 11.84CALAVLLLFLNLPAYY30 pKa = 8.54PINEE34 pKa = 3.81ICKK37 pKa = 10.34AMDD40 pKa = 4.91SIPEE44 pKa = 4.1SDD46 pKa = 3.11SMARR50 pKa = 11.84IFAAMDD56 pKa = 3.17VFDD59 pKa = 5.6NYY61 pKa = 10.86TGDD64 pKa = 3.89LACFDD69 pKa = 4.92IYY71 pKa = 11.23DD72 pKa = 4.82DD73 pKa = 3.79PHH75 pKa = 8.72GMSGWDD81 pKa = 3.08
MM1 pKa = 7.39AQTEE5 pKa = 4.19LALPSGRR12 pKa = 11.84LQRR15 pKa = 11.84CALAVLLLFLNLPAYY30 pKa = 8.54PINEE34 pKa = 3.81ICKK37 pKa = 10.34AMDD40 pKa = 4.91SIPEE44 pKa = 4.1SDD46 pKa = 3.11SMARR50 pKa = 11.84IFAAMDD56 pKa = 3.17VFDD59 pKa = 5.6NYY61 pKa = 10.86TGDD64 pKa = 3.89LACFDD69 pKa = 4.92IYY71 pKa = 11.23DD72 pKa = 4.82DD73 pKa = 3.79PHH75 pKa = 8.72GMSGWDD81 pKa = 3.08
Molecular weight: 8.96 kDa
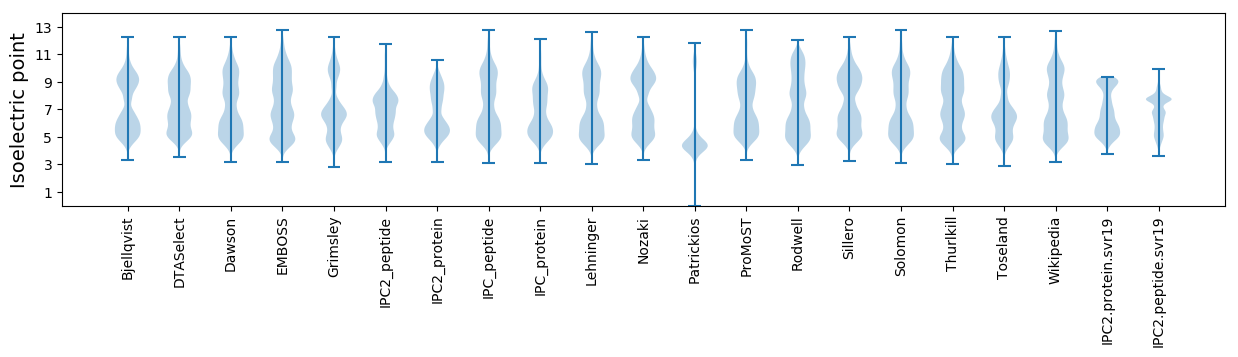
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A176VRM0|A0A176VRM0_MARPO Uncharacterized protein OS=Marchantia polymorpha subsp. ruderalis OX=1480154 GN=AXG93_285s1370 PE=4 SV=1
MM1 pKa = 7.37TMYY4 pKa = 10.85DD5 pKa = 3.88DD6 pKa = 4.69SDD8 pKa = 3.71PTRR11 pKa = 11.84GMLGHH16 pKa = 6.99GGMGMGASMRR26 pKa = 11.84HH27 pKa = 5.28TGQPSTPVFRR37 pKa = 11.84VVGPIPACGYY47 pKa = 10.35APEE50 pKa = 4.58FNTALSHH57 pKa = 6.1SPSPKK62 pKa = 8.29STLSRR67 pKa = 11.84LHH69 pKa = 6.87DD70 pKa = 4.02FARR73 pKa = 11.84RR74 pKa = 11.84NYY76 pKa = 7.87KK77 pKa = 8.63TRR79 pKa = 11.84SRR81 pKa = 11.84SRR83 pKa = 11.84LVSALLIVTEE93 pKa = 4.02QARR96 pKa = 11.84ACASAWACARR106 pKa = 11.84AQPQAPEE113 pKa = 4.08SQRR116 pKa = 11.84TRR118 pKa = 11.84MRR120 pKa = 11.84YY121 pKa = 9.33FLGKK125 pKa = 9.23PWGQRR130 pKa = 11.84KK131 pKa = 7.74YY132 pKa = 10.35VRR134 pKa = 11.84RR135 pKa = 11.84DD136 pKa = 2.82GNAEE140 pKa = 3.82HH141 pKa = 7.14GSGKK145 pKa = 10.22KK146 pKa = 9.84SPAWRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84KK154 pKa = 8.57VTAAGEE160 pKa = 4.2SRR162 pKa = 11.84AGDD165 pKa = 3.59EE166 pKa = 4.34TVSAVTGEE174 pKa = 4.29SVRR177 pKa = 11.84NSGVTVPTTEE187 pKa = 4.59PLPLDD192 pKa = 3.65LTFLISPQGQNPWLIPSRR210 pKa = 11.84ALDD213 pKa = 3.49GNARR217 pKa = 11.84RR218 pKa = 11.84EE219 pKa = 4.31LRR221 pKa = 11.84CSPLPHH227 pKa = 6.79GSSEE231 pKa = 4.05PAVAEE236 pKa = 4.16LTCGFGVGPLKK247 pKa = 10.56FGHH250 pKa = 7.15DD251 pKa = 4.26NFPVILPRR259 pKa = 11.84VRR261 pKa = 3.77
MM1 pKa = 7.37TMYY4 pKa = 10.85DD5 pKa = 3.88DD6 pKa = 4.69SDD8 pKa = 3.71PTRR11 pKa = 11.84GMLGHH16 pKa = 6.99GGMGMGASMRR26 pKa = 11.84HH27 pKa = 5.28TGQPSTPVFRR37 pKa = 11.84VVGPIPACGYY47 pKa = 10.35APEE50 pKa = 4.58FNTALSHH57 pKa = 6.1SPSPKK62 pKa = 8.29STLSRR67 pKa = 11.84LHH69 pKa = 6.87DD70 pKa = 4.02FARR73 pKa = 11.84RR74 pKa = 11.84NYY76 pKa = 7.87KK77 pKa = 8.63TRR79 pKa = 11.84SRR81 pKa = 11.84SRR83 pKa = 11.84LVSALLIVTEE93 pKa = 4.02QARR96 pKa = 11.84ACASAWACARR106 pKa = 11.84AQPQAPEE113 pKa = 4.08SQRR116 pKa = 11.84TRR118 pKa = 11.84MRR120 pKa = 11.84YY121 pKa = 9.33FLGKK125 pKa = 9.23PWGQRR130 pKa = 11.84KK131 pKa = 7.74YY132 pKa = 10.35VRR134 pKa = 11.84RR135 pKa = 11.84DD136 pKa = 2.82GNAEE140 pKa = 3.82HH141 pKa = 7.14GSGKK145 pKa = 10.22KK146 pKa = 9.84SPAWRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84KK154 pKa = 8.57VTAAGEE160 pKa = 4.2SRR162 pKa = 11.84AGDD165 pKa = 3.59EE166 pKa = 4.34TVSAVTGEE174 pKa = 4.29SVRR177 pKa = 11.84NSGVTVPTTEE187 pKa = 4.59PLPLDD192 pKa = 3.65LTFLISPQGQNPWLIPSRR210 pKa = 11.84ALDD213 pKa = 3.49GNARR217 pKa = 11.84RR218 pKa = 11.84EE219 pKa = 4.31LRR221 pKa = 11.84CSPLPHH227 pKa = 6.79GSSEE231 pKa = 4.05PAVAEE236 pKa = 4.16LTCGFGVGPLKK247 pKa = 10.56FGHH250 pKa = 7.15DD251 pKa = 4.26NFPVILPRR259 pKa = 11.84VRR261 pKa = 3.77
Molecular weight: 28.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7780001 |
66 |
7651 |
433.4 |
47.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.302 ± 0.018 | 1.656 ± 0.008 |
5.344 ± 0.012 | 6.903 ± 0.022 |
3.552 ± 0.012 | 7.233 ± 0.019 |
2.226 ± 0.008 | 4.236 ± 0.012 |
5.335 ± 0.018 | 9.397 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.293 ± 0.007 | 3.446 ± 0.01 |
5.198 ± 0.021 | 3.957 ± 0.014 |
6.457 ± 0.017 | 8.69 ± 0.022 |
5.14 ± 0.011 | 6.927 ± 0.014 |
1.334 ± 0.007 | 2.361 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
