
Actinokineospora auranticolor
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinokineospora
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
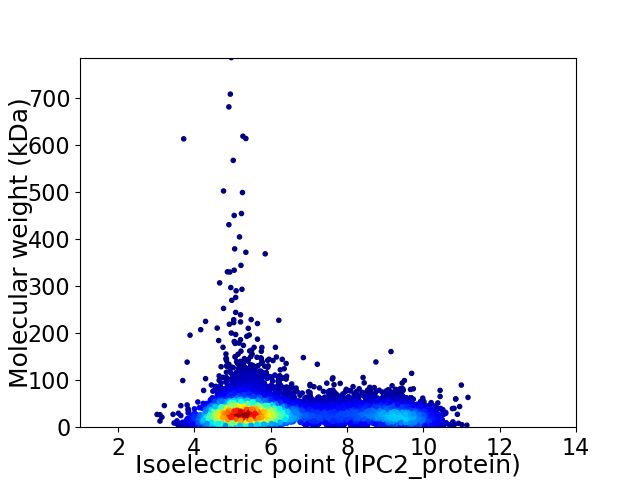
Virtual 2D-PAGE plot for 7400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S6GTR1|A0A2S6GTR1_9PSEU Phosphotransferase family enzyme OS=Actinokineospora auranticolor OX=155976 GN=CLV40_105308 PE=4 SV=1
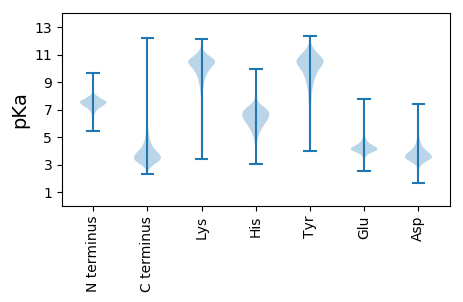
MM1 pKa = 7.61TEE3 pKa = 4.45PDD5 pKa = 3.67KK6 pKa = 11.26TWDD9 pKa = 3.74DD10 pKa = 3.32GHH12 pKa = 7.9YY13 pKa = 11.22GEE15 pKa = 6.08DD16 pKa = 4.08DD17 pKa = 3.96VIEE20 pKa = 4.26AADD23 pKa = 3.65GKK25 pKa = 11.17GYY27 pKa = 10.79LSVSDD32 pKa = 4.1YY33 pKa = 10.79THH35 pKa = 7.08GDD37 pKa = 3.24NGLEE41 pKa = 4.29PDD43 pKa = 3.57DD44 pKa = 5.17HH45 pKa = 7.64SWW47 pKa = 3.05
MM1 pKa = 7.61TEE3 pKa = 4.45PDD5 pKa = 3.67KK6 pKa = 11.26TWDD9 pKa = 3.74DD10 pKa = 3.32GHH12 pKa = 7.9YY13 pKa = 11.22GEE15 pKa = 6.08DD16 pKa = 4.08DD17 pKa = 3.96VIEE20 pKa = 4.26AADD23 pKa = 3.65GKK25 pKa = 11.17GYY27 pKa = 10.79LSVSDD32 pKa = 4.1YY33 pKa = 10.79THH35 pKa = 7.08GDD37 pKa = 3.24NGLEE41 pKa = 4.29PDD43 pKa = 3.57DD44 pKa = 5.17HH45 pKa = 7.64SWW47 pKa = 3.05
Molecular weight: 5.24 kDa
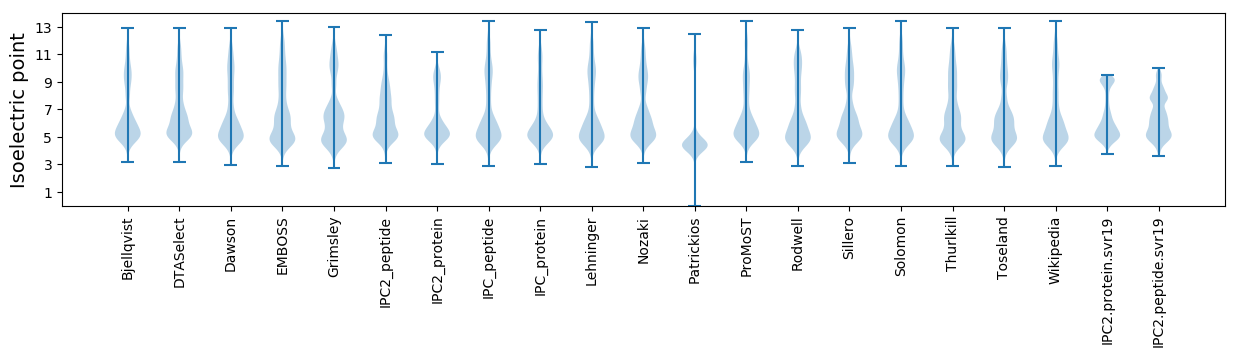
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S6GZ32|A0A2S6GZ32_9PSEU Betaine-aldehyde dehydrogenase OS=Actinokineospora auranticolor OX=155976 GN=CLV40_102341 PE=3 SV=1
MM1 pKa = 7.66AWPARR6 pKa = 11.84PPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PAGRR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84TPPRR22 pKa = 11.84RR23 pKa = 11.84PGAPTSRR30 pKa = 11.84TRR32 pKa = 11.84PPSASPATAWASPRR46 pKa = 11.84PAPPGARR53 pKa = 11.84APPLPAPPGRR63 pKa = 11.84PRR65 pKa = 11.84PHH67 pKa = 6.76RR68 pKa = 11.84AAPRR72 pKa = 11.84PPQARR77 pKa = 11.84QTPRR81 pKa = 11.84PHH83 pKa = 6.69VPTPQGPAHH92 pKa = 6.86HH93 pKa = 7.02AHH95 pKa = 5.84QACPTARR102 pKa = 11.84TLRR105 pKa = 11.84VRR107 pKa = 11.84GRR109 pKa = 11.84PRR111 pKa = 11.84VRR113 pKa = 11.84RR114 pKa = 11.84GPRR117 pKa = 11.84PRR119 pKa = 11.84PVTGALQPRR128 pKa = 11.84VQVRR132 pKa = 11.84LRR134 pKa = 11.84ARR136 pKa = 11.84PVRR139 pKa = 11.84PLLPRR144 pKa = 11.84HH145 pKa = 5.85RR146 pKa = 11.84RR147 pKa = 11.84PPRR150 pKa = 11.84RR151 pKa = 11.84PARR154 pKa = 11.84ARR156 pKa = 11.84QGPARR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84LAGRR168 pKa = 11.84ARR170 pKa = 11.84LRR172 pKa = 11.84ARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84GRR178 pKa = 11.84PPGARR183 pKa = 11.84VGRR186 pKa = 11.84LPRR189 pKa = 11.84PGRR192 pKa = 11.84GSPATHH198 pKa = 6.34HH199 pKa = 7.03LSRR202 pKa = 11.84HH203 pKa = 4.92LVGSLRR209 pKa = 11.84RR210 pKa = 11.84DD211 pKa = 3.2LARR214 pKa = 11.84PGKK217 pKa = 7.8TAAPRR222 pKa = 11.84PRR224 pKa = 11.84DD225 pKa = 3.05RR226 pKa = 11.84ARR228 pKa = 11.84RR229 pKa = 11.84EE230 pKa = 3.63RR231 pKa = 11.84VRR233 pKa = 11.84PHH235 pKa = 5.83PSKK238 pKa = 10.19RR239 pKa = 11.84TAHH242 pKa = 7.2RR243 pKa = 11.84SRR245 pKa = 11.84VCTGSSARR253 pKa = 11.84RR254 pKa = 11.84TRR256 pKa = 11.84HH257 pKa = 5.87RR258 pKa = 11.84GRR260 pKa = 11.84IDD262 pKa = 3.22RR263 pKa = 11.84VRR265 pKa = 11.84GKK267 pKa = 9.45VRR269 pKa = 11.84RR270 pKa = 11.84SRR272 pKa = 11.84TAPGPTRR279 pKa = 11.84DD280 pKa = 3.67TAHH283 pKa = 6.7KK284 pKa = 8.51VHH286 pKa = 7.3RR287 pKa = 11.84SRR289 pKa = 11.84ASRR292 pKa = 11.84SGVGTSRR299 pKa = 11.84IRR301 pKa = 11.84ATVRR305 pKa = 11.84GRR307 pKa = 11.84RR308 pKa = 11.84QAHH311 pKa = 6.32RR312 pKa = 11.84PSQATAGHH320 pKa = 7.03RR321 pKa = 11.84ANGHH325 pKa = 5.58RR326 pKa = 11.84PCPSHH331 pKa = 5.79MALRR335 pKa = 11.84PGLVTLRR342 pKa = 11.84RR343 pKa = 11.84RR344 pKa = 11.84ANAPRR349 pKa = 11.84VRR351 pKa = 11.84RR352 pKa = 11.84PAARR356 pKa = 11.84LSRR359 pKa = 11.84TALRR363 pKa = 11.84PKK365 pKa = 10.64GLTLRR370 pKa = 11.84RR371 pKa = 11.84NAPRR375 pKa = 11.84ARR377 pKa = 11.84RR378 pKa = 11.84TAPHH382 pKa = 7.18PEE384 pKa = 3.49RR385 pKa = 11.84ATRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84AMVSPRR397 pKa = 11.84RR398 pKa = 11.84VIARR402 pKa = 11.84SRR404 pKa = 11.84ALDD407 pKa = 3.38RR408 pKa = 11.84PTRR411 pKa = 11.84GRR413 pKa = 11.84RR414 pKa = 11.84PPPVRR419 pKa = 11.84HH420 pKa = 6.41DD421 pKa = 3.5RR422 pKa = 11.84APVRR426 pKa = 11.84TGNRR430 pKa = 11.84RR431 pKa = 11.84PDD433 pKa = 3.23RR434 pKa = 11.84PRR436 pKa = 11.84GLRR439 pKa = 11.84MATVPTGTTRR449 pKa = 11.84RR450 pKa = 11.84RR451 pKa = 11.84GASRR455 pKa = 11.84QRR457 pKa = 11.84RR458 pKa = 11.84TRR460 pKa = 11.84GPNHH464 pKa = 5.9VVRR467 pKa = 11.84TTPGARR473 pKa = 11.84SRR475 pKa = 11.84RR476 pKa = 11.84EE477 pKa = 3.75TSATAPTRR485 pKa = 11.84TPPANAPATTRR496 pKa = 11.84AGARR500 pKa = 11.84TRR502 pKa = 11.84TQPTTGPRR510 pKa = 11.84TRR512 pKa = 11.84TRR514 pKa = 11.84PRR516 pKa = 11.84ITGGTRR522 pKa = 11.84TGPSRR527 pKa = 11.84TGRR530 pKa = 11.84PTRR533 pKa = 11.84GSRR536 pKa = 11.84PTSAGTPRR544 pKa = 11.84TSRR547 pKa = 11.84TSTTPPTPSCWCARR561 pKa = 11.84STARR565 pKa = 11.84RR566 pKa = 11.84RR567 pKa = 11.84AAA569 pKa = 3.47
MM1 pKa = 7.66AWPARR6 pKa = 11.84PPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PAGRR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84TPPRR22 pKa = 11.84RR23 pKa = 11.84PGAPTSRR30 pKa = 11.84TRR32 pKa = 11.84PPSASPATAWASPRR46 pKa = 11.84PAPPGARR53 pKa = 11.84APPLPAPPGRR63 pKa = 11.84PRR65 pKa = 11.84PHH67 pKa = 6.76RR68 pKa = 11.84AAPRR72 pKa = 11.84PPQARR77 pKa = 11.84QTPRR81 pKa = 11.84PHH83 pKa = 6.69VPTPQGPAHH92 pKa = 6.86HH93 pKa = 7.02AHH95 pKa = 5.84QACPTARR102 pKa = 11.84TLRR105 pKa = 11.84VRR107 pKa = 11.84GRR109 pKa = 11.84PRR111 pKa = 11.84VRR113 pKa = 11.84RR114 pKa = 11.84GPRR117 pKa = 11.84PRR119 pKa = 11.84PVTGALQPRR128 pKa = 11.84VQVRR132 pKa = 11.84LRR134 pKa = 11.84ARR136 pKa = 11.84PVRR139 pKa = 11.84PLLPRR144 pKa = 11.84HH145 pKa = 5.85RR146 pKa = 11.84RR147 pKa = 11.84PPRR150 pKa = 11.84RR151 pKa = 11.84PARR154 pKa = 11.84ARR156 pKa = 11.84QGPARR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84LAGRR168 pKa = 11.84ARR170 pKa = 11.84LRR172 pKa = 11.84ARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84GRR178 pKa = 11.84PPGARR183 pKa = 11.84VGRR186 pKa = 11.84LPRR189 pKa = 11.84PGRR192 pKa = 11.84GSPATHH198 pKa = 6.34HH199 pKa = 7.03LSRR202 pKa = 11.84HH203 pKa = 4.92LVGSLRR209 pKa = 11.84RR210 pKa = 11.84DD211 pKa = 3.2LARR214 pKa = 11.84PGKK217 pKa = 7.8TAAPRR222 pKa = 11.84PRR224 pKa = 11.84DD225 pKa = 3.05RR226 pKa = 11.84ARR228 pKa = 11.84RR229 pKa = 11.84EE230 pKa = 3.63RR231 pKa = 11.84VRR233 pKa = 11.84PHH235 pKa = 5.83PSKK238 pKa = 10.19RR239 pKa = 11.84TAHH242 pKa = 7.2RR243 pKa = 11.84SRR245 pKa = 11.84VCTGSSARR253 pKa = 11.84RR254 pKa = 11.84TRR256 pKa = 11.84HH257 pKa = 5.87RR258 pKa = 11.84GRR260 pKa = 11.84IDD262 pKa = 3.22RR263 pKa = 11.84VRR265 pKa = 11.84GKK267 pKa = 9.45VRR269 pKa = 11.84RR270 pKa = 11.84SRR272 pKa = 11.84TAPGPTRR279 pKa = 11.84DD280 pKa = 3.67TAHH283 pKa = 6.7KK284 pKa = 8.51VHH286 pKa = 7.3RR287 pKa = 11.84SRR289 pKa = 11.84ASRR292 pKa = 11.84SGVGTSRR299 pKa = 11.84IRR301 pKa = 11.84ATVRR305 pKa = 11.84GRR307 pKa = 11.84RR308 pKa = 11.84QAHH311 pKa = 6.32RR312 pKa = 11.84PSQATAGHH320 pKa = 7.03RR321 pKa = 11.84ANGHH325 pKa = 5.58RR326 pKa = 11.84PCPSHH331 pKa = 5.79MALRR335 pKa = 11.84PGLVTLRR342 pKa = 11.84RR343 pKa = 11.84RR344 pKa = 11.84ANAPRR349 pKa = 11.84VRR351 pKa = 11.84RR352 pKa = 11.84PAARR356 pKa = 11.84LSRR359 pKa = 11.84TALRR363 pKa = 11.84PKK365 pKa = 10.64GLTLRR370 pKa = 11.84RR371 pKa = 11.84NAPRR375 pKa = 11.84ARR377 pKa = 11.84RR378 pKa = 11.84TAPHH382 pKa = 7.18PEE384 pKa = 3.49RR385 pKa = 11.84ATRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84AMVSPRR397 pKa = 11.84RR398 pKa = 11.84VIARR402 pKa = 11.84SRR404 pKa = 11.84ALDD407 pKa = 3.38RR408 pKa = 11.84PTRR411 pKa = 11.84GRR413 pKa = 11.84RR414 pKa = 11.84PPPVRR419 pKa = 11.84HH420 pKa = 6.41DD421 pKa = 3.5RR422 pKa = 11.84APVRR426 pKa = 11.84TGNRR430 pKa = 11.84RR431 pKa = 11.84PDD433 pKa = 3.23RR434 pKa = 11.84PRR436 pKa = 11.84GLRR439 pKa = 11.84MATVPTGTTRR449 pKa = 11.84RR450 pKa = 11.84RR451 pKa = 11.84GASRR455 pKa = 11.84QRR457 pKa = 11.84RR458 pKa = 11.84TRR460 pKa = 11.84GPNHH464 pKa = 5.9VVRR467 pKa = 11.84TTPGARR473 pKa = 11.84SRR475 pKa = 11.84RR476 pKa = 11.84EE477 pKa = 3.75TSATAPTRR485 pKa = 11.84TPPANAPATTRR496 pKa = 11.84AGARR500 pKa = 11.84TRR502 pKa = 11.84TQPTTGPRR510 pKa = 11.84TRR512 pKa = 11.84TRR514 pKa = 11.84PRR516 pKa = 11.84ITGGTRR522 pKa = 11.84TGPSRR527 pKa = 11.84TGRR530 pKa = 11.84PTRR533 pKa = 11.84GSRR536 pKa = 11.84PTSAGTPRR544 pKa = 11.84TSRR547 pKa = 11.84TSTTPPTPSCWCARR561 pKa = 11.84STARR565 pKa = 11.84RR566 pKa = 11.84RR567 pKa = 11.84AAA569 pKa = 3.47
Molecular weight: 63.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2468596 |
26 |
7590 |
333.6 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.79 ± 0.045 | 0.777 ± 0.009 |
6.312 ± 0.026 | 5.196 ± 0.03 |
2.714 ± 0.014 | 9.158 ± 0.03 |
2.277 ± 0.015 | 3.051 ± 0.018 |
1.8 ± 0.024 | 10.406 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.483 ± 0.012 | 1.816 ± 0.021 |
6.227 ± 0.026 | 2.492 ± 0.017 |
8.305 ± 0.04 | 4.901 ± 0.026 |
6.488 ± 0.042 | 9.406 ± 0.033 |
1.524 ± 0.012 | 1.875 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
